上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
View or download extensive performance data in our Data Supplement.
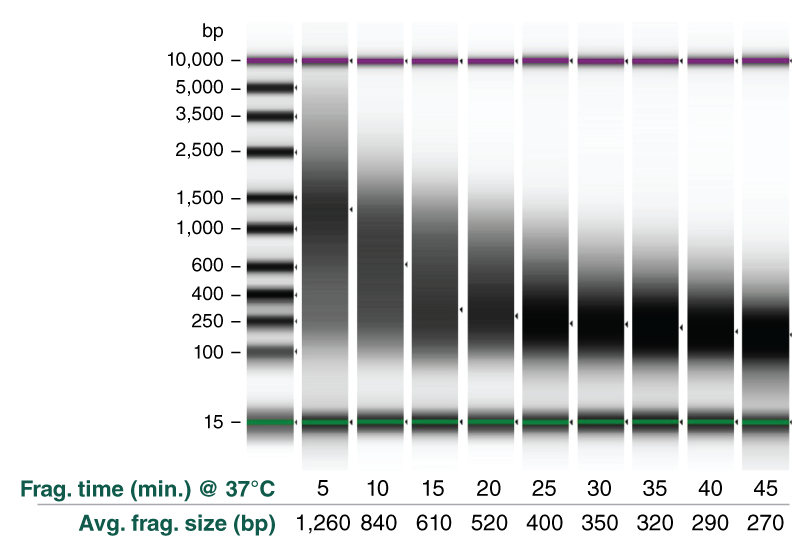
Enzymatic methods for DNA fragmentation in NGS workflows enable streamlined protocols, improved performance and scalability. However, specialized fragmentation reagents are required for samples for methylation analysis, to ensure that methylation marks are not removed, and for FFPE DNA. NEBNext UltraShear is a novel enzyme mix designed for fragmentation of these sample types that has a fast workflow and improves library preparation and sequencing metrics for DNA methylation studies and FFPE DNA.
NEBNext UltraShear is compatible with NEBNext Enzymatic Methyl-seq (EM-seq) (NEB #E7120).
NEBNext UltraShear in FFPE DNA sequencing workflows:
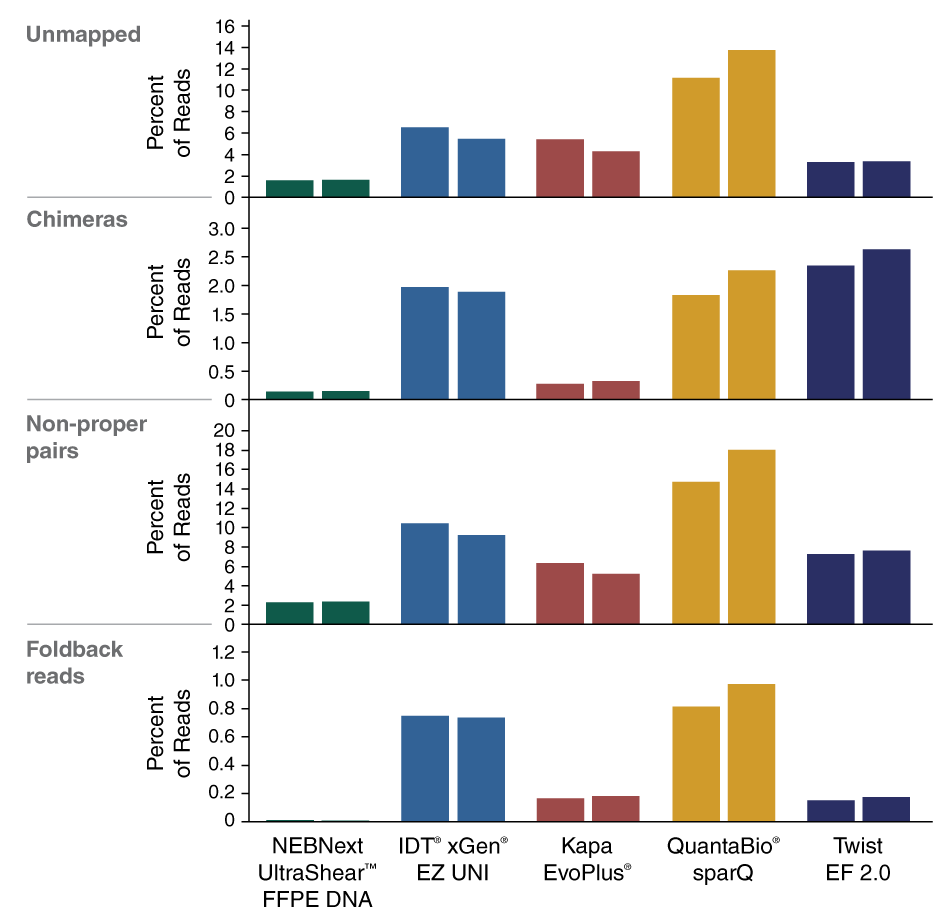
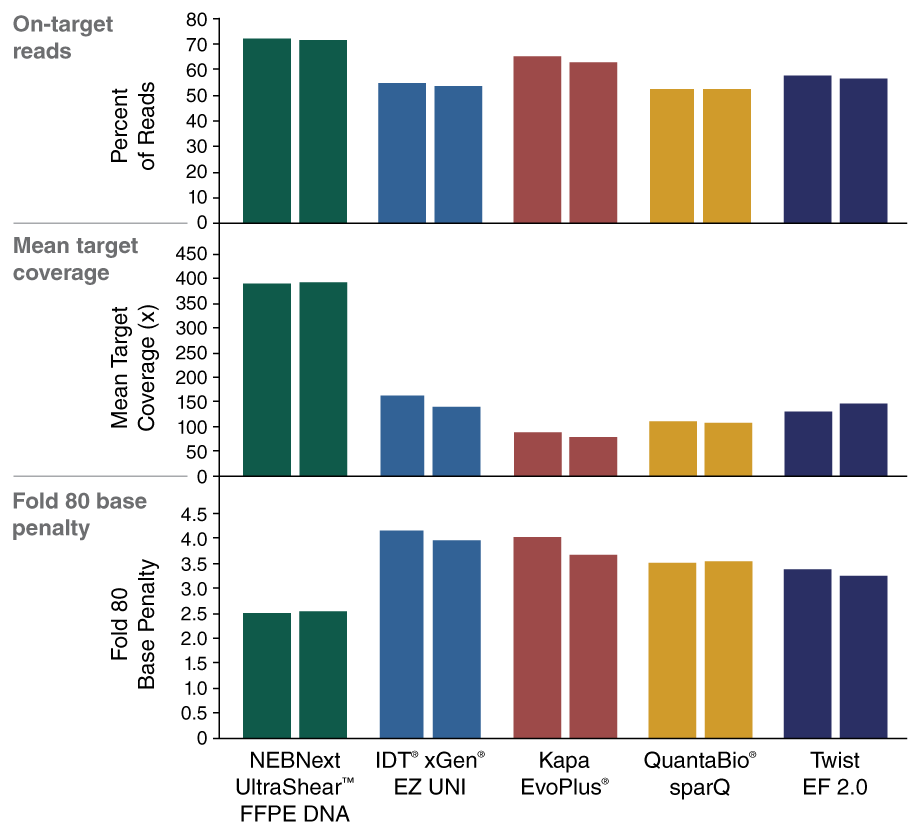
- Improved usable reads
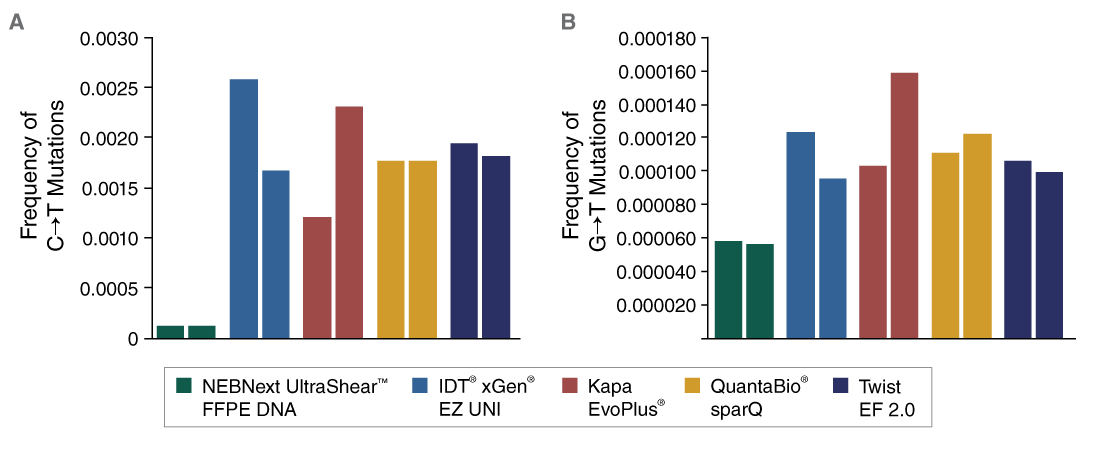
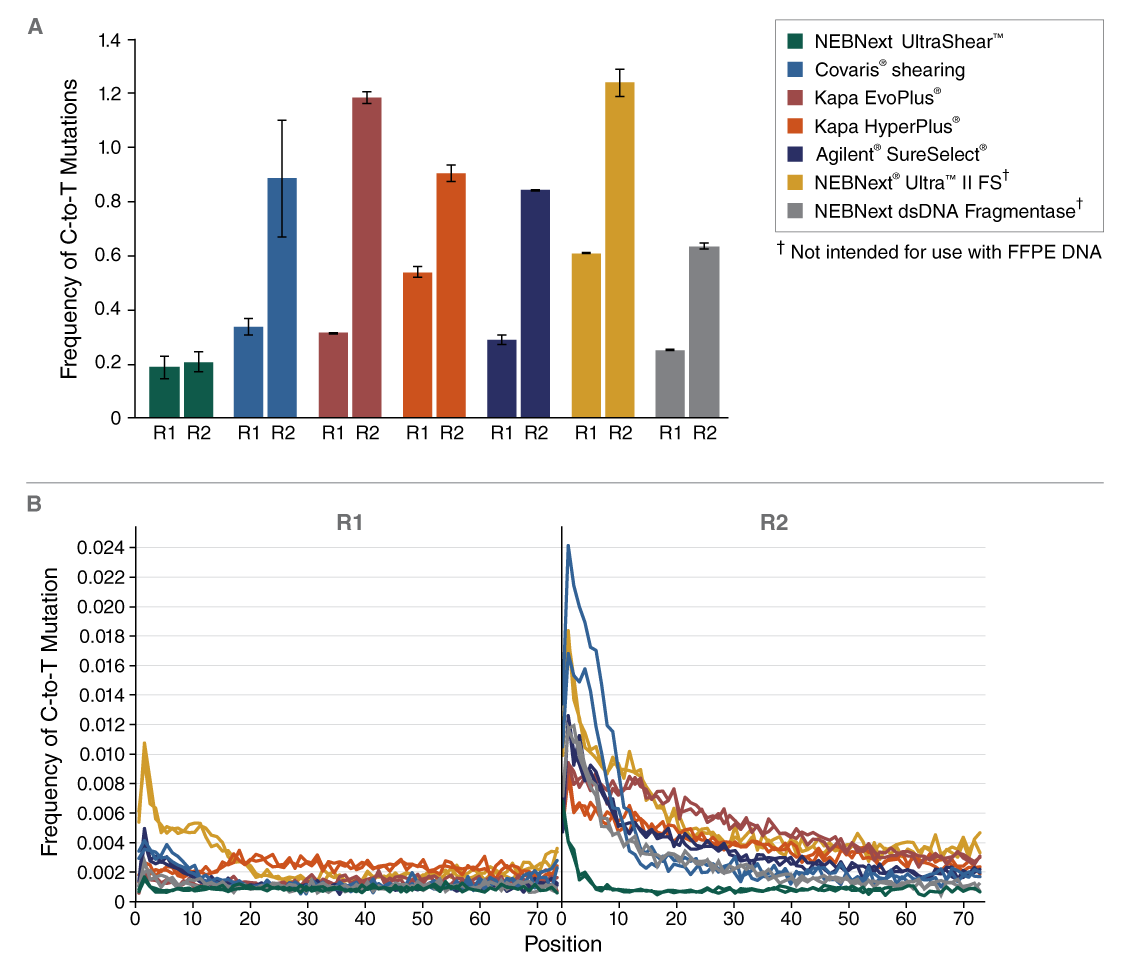
- Lower artificial mutation frequency
NEBNext UltraShear for methylated DNA sequencing e.g., EM-seq
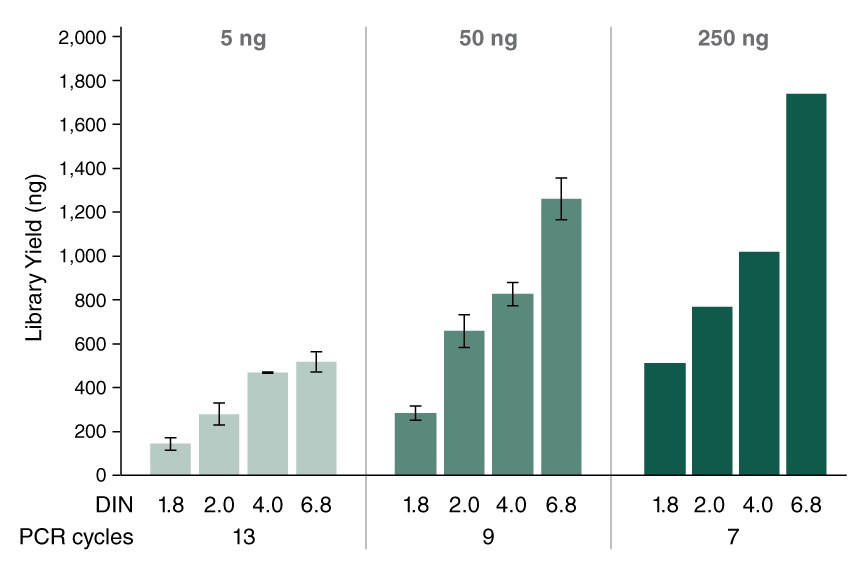
- Higher library yields
- Improved sequencing metrics
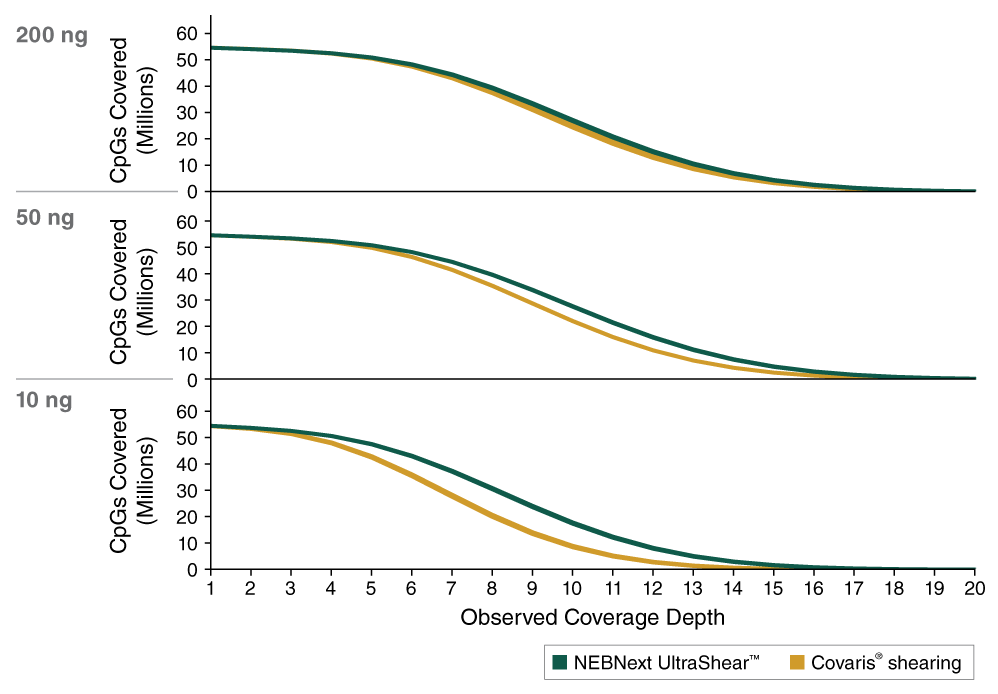
- Improved CpG Coverage
Note that for high quality genomic DNA library prep with enzymatic fragmentation, we recommend NEBNext Ultra II FS DNA Library Prep for Illumina (NEB #E7805, #E6177).
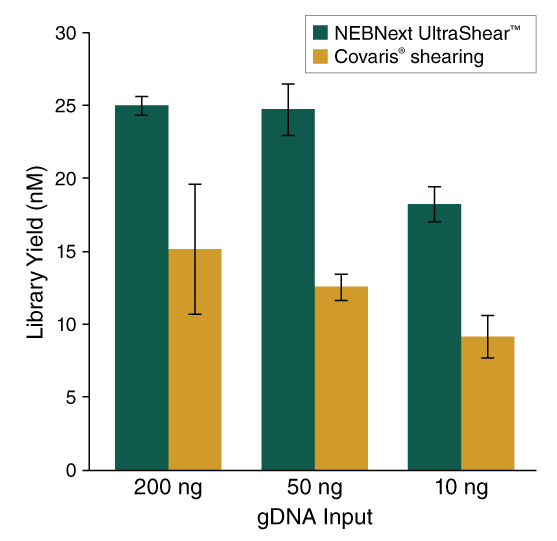
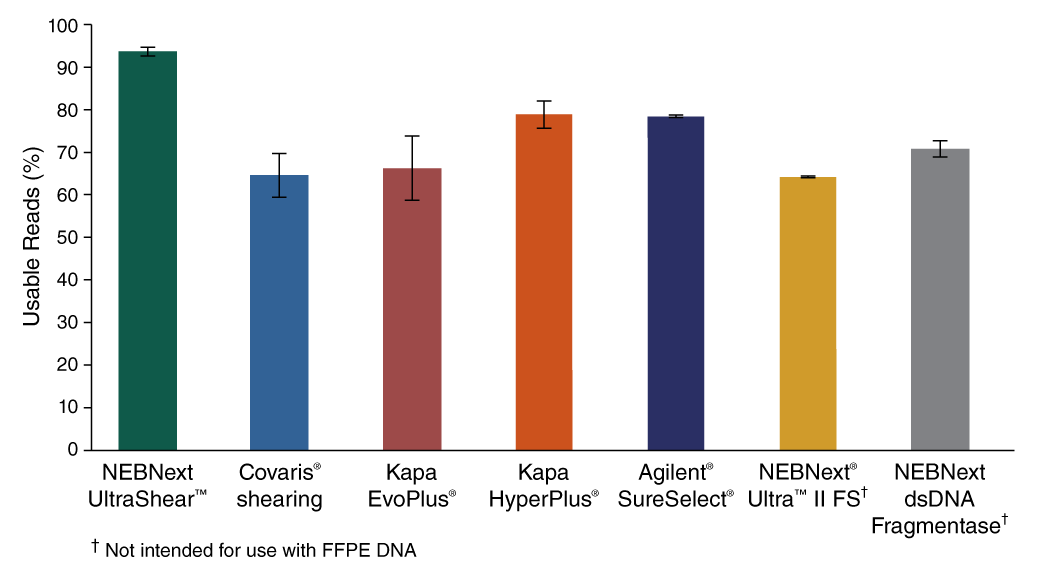
- 产品类别:
- FFPE DNA Products,
- DNA Fragmentation & RNA Fragmentation Products,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
M7634S -20 NEBNext UltraShear® M7634SVIAL -20 1 x 0.096 ml Not Applicable NEBNext UltraShear™ Reaction Buffer B9042SVIAL -20 1 x 0.336 ml Not Applicable 500 mM DTT B1079SVIAL -20 1 x 0.048 ml Not Applicable
-
M7634L -20 NEBNext UltraShear® M7634LVIAL -20 1 x 0.384 ml Not Applicable NEBNext UltraShear™ Reaction Buffer B9042LVIAL -20 2 x 0.672 ml Not Applicable 500 mM DTT B1079LVIAL -20 1 x 0.192 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
- 1X TE (10 mM Tris pH 8.0, 1 mM EDTA)
- 0.2 ml thin wall PCR tubes
- Magnetic rack/stand (NEB #S1515S; Alpaqua® #A001322 or equivalent)
- PCR machine
- Vortex
- Microcentrifuge
- Bioanalyzer®, TapeStation® or other fragment analyzer and associated consumables
- 80% Ethanol
For use with NEBNext UltraShear Protocol:
- SPRIselect™ Reagent Kit (Beckman Coulter, Inc. #B23317), AMPure® XP Beads (Beckman Coulter, Inc. #A63881) or Monarch®® PCR & DNA Cleanup Kit (NEB# T1030S/L) are recommended for Section 1.
For use with NEBNext Ultra II End Repair/dA-Tailing Module Protocol:
- NEBNext Ultra II End Repair/dA-Tailing Module (NEB #E7546S/L) for Section 2.
- Recommended Material Not Included: NEBNext Ultra II Ligation Module (NEB #E7595) and NEBNext Multiplex Oligos (www.neb.com/oligos).
For use with NEBNext Enzymatic Methyl-seq Protocol:
- NEBNext Enzymatic Methyl-seq Kit (NEB #E7120S/L) for Section 3.
- Formamide (Sigma #F9037-100 ml) or optional 0.1 N NaOH. Formamide is preferred. If using NaOH, please see NEBNext Enzymatic Methyl-seq Kit (NEB #E7120) FAQs
- Nuclease-free
-
相关产品
相关产品
- NEBNext® 酶学转化法甲基化建库试剂盒
- NEBNext® 甲基化建库酶学转化法模块
- E7360 NEBNext FFPE DNA 修复模块 v2
操作说明、说明书 & 用法
-
操作说明
- Where can I find guidelines and protocols for using NEBNext UltraShear, including in conjunction with NEBNext Enzymatic Methyl-seq (EM-seq™)?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualM7634
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- Is NEBNext UltraShear™ the same as NEBNext® Ultra™ II FS or NEBNext dsDNA Fragmentase®?
- Do you really need to vortex NEBNext UltraShear™?
- For EM-seq™ workflows, what are the recommendations for fragmenting already-fragmented DNA, low integrity DNA and/or FFPE DNA with NEBNext UltraShear™?
- Following the fragmentation step of the NEBNext UltraShear™ protocol, can the reactions be stored at -20˚C?
- Do you recommend NEBNext UltraShear™ for high-sensitivity, low error rate DNA library preparation?