上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
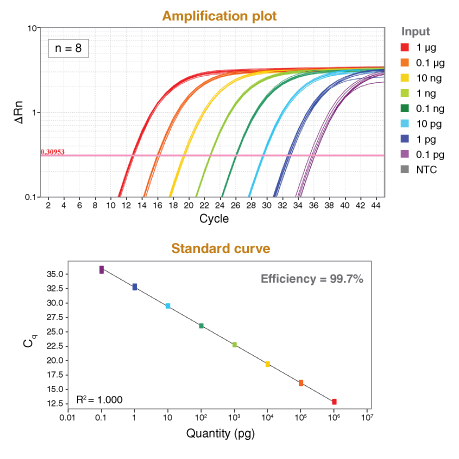
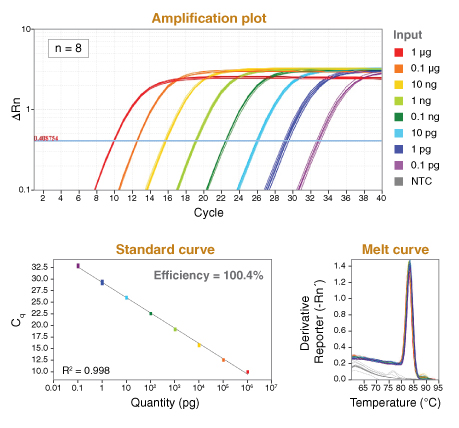
探针法定量 PCR(qPCR)原理是利用聚合酶的 5´→3´ 核酸外切酶活性,将淬灭的靶基因特异性探针切断,使其发出荧光,并实时监测释放出的荧光,以测量每个 PCR 循环中的 DNA 扩增。通过检测荧光信号显著超过本底信号时的循环数,可确定 Cq 值。Cq 值可用于评估评估两个或两个以上的样本之间的相对丰度,也通过梯度稀释已知浓度样品所绘制的标准曲线,对样本绝对定量。
Luna 通用探针法 qPCR 预混液是经优化的 2X 反应预混液,适用于水解探针法对靶标 DNA 序列进行实时 qPCR 检测和定量分析。该预混液包含热启动 Taq DNA 聚合酶,还添加了独特的惰性参比染料,该染料可与多种 qPCR 仪器兼容(包括需要高 ROX 或低 ROX 参比信号的仪器)。独具特色的是:预混液还包含可防止交叉污染的 dUTP,以及有助于观察反应体系建立的非荧光可见示踪染料。该染料的光谱与 qPCR 荧光染料不重叠,因此不会影响到实时检测数值。
该预混液浓度为 2X,包含 DNA 扩增和定量所需的所有 PCR 组分(引物/探针和 DNA 模板除外)。使用 Luna qPCR 试剂配合商品化的 qPCR 检测引物/探针,可以对基因组 DNA 或感兴趣的 cDNA 进行定量。
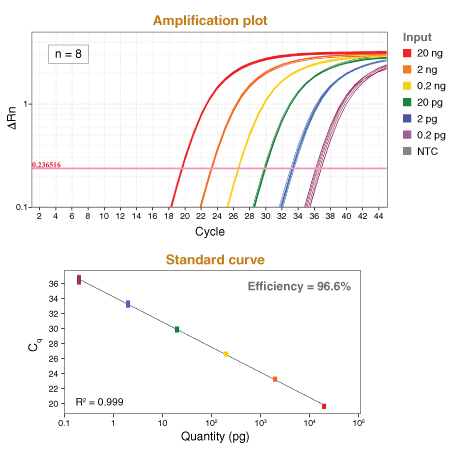
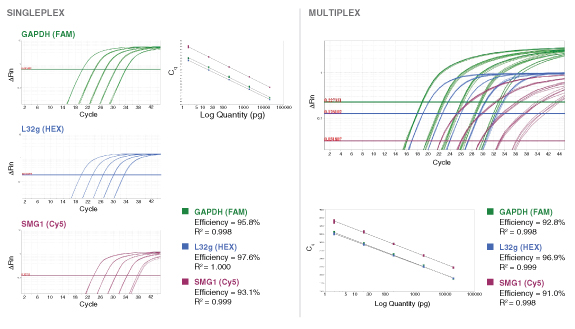
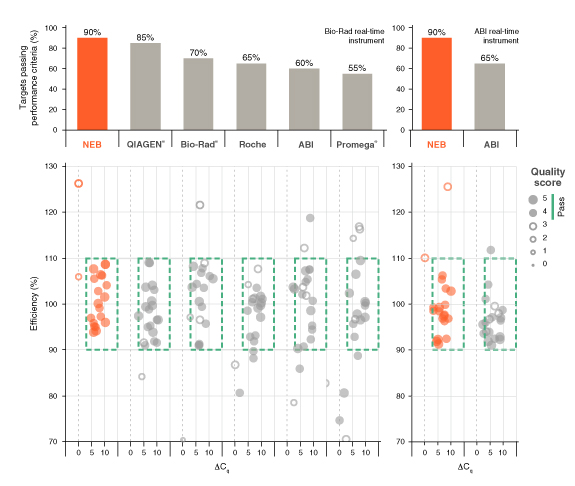
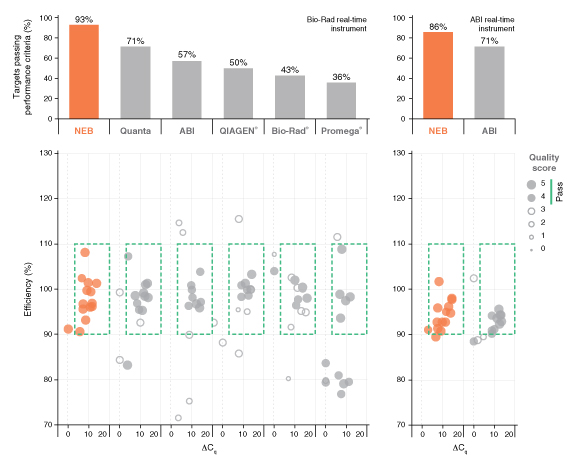
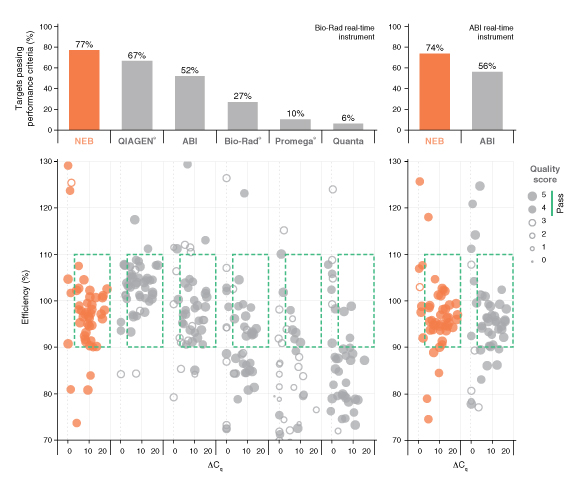
详细了解 qPCR/RT-qPCR 综合测试和“dots in boxes”数据可视化分析方法

- 产品类别:
- Luna® qPCR & RT-qPCR Products,
- PCR, qPCR & Amplification Technologies Products
- 应用:
- qPCR & RT-qPCR,
- DNA Amplification, PCR & qPCR
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
M3004V -20 Luna® Universal Probe qPCR Master Mix M3004VVIAL -20 1 x 1 ml 2 X
-
M3004S -20 Luna® Universal Probe qPCR Master Mix M3004SVIAL -20 2 x 1 ml 2 X
-
M3004L -20 Luna® Universal Probe qPCR Master Mix M3004SVIAL -20 5 x 1 ml 2 X
-
M3004X -20 Luna® Universal Probe qPCR Master Mix M3004SVIAL -20 10 x 1 ml 2 X
-
M3004E -20 Luna® Universal Probe qPCR Master Mix M3004EVIAL -20 1 x 25 ml 2 X
-
-
相关产品
相关产品
- 南极热敏 UDG
- Luna® 通用 qPCR 预混液
- Luna® 探针一步法 RT-qPCR 试剂盒(无 ROX)
- Luna® 通用一步法 RT-qPCR 试剂盒
- Luna® 通用探针一步法 RT-qPCR 试剂盒
- LunaScript™ 反转录 SuperMix 试剂盒
-
注意事项
- Assay Design
The use of qPCR primer design software (e.g., Primer3) maximizes the likelihood of amplification success while minimizing nonspecific amplification and primer dimers. Targets with balanced GC/AT content (40–60%) tend to amplify efficiently. Where possible, enter sufficient sequence around the area of interest to permit robust primer design and use search criteria that permit cross-reference against relevant sequence databases (to avoid potential off-target amplification). For cDNA targets, it is advisable to design primers across known splicing sites in order to prevent amplification from genomic DNA. Conversely, primers designed to target intronic regions can ensure amplification exclusively from genomic DNA. - Primer and Probe Concentration
For most targets, a final concentration of 400 nM for each primer will provide optimum performance. If needed, primer concentrations can be optimized between 200–900 nM. Probe should be included at 200 nM for best results. Probe concentration can be optimized in the range of 100–500 nM if optimization of performance or target fluorescence level is desired. - Multiplexing
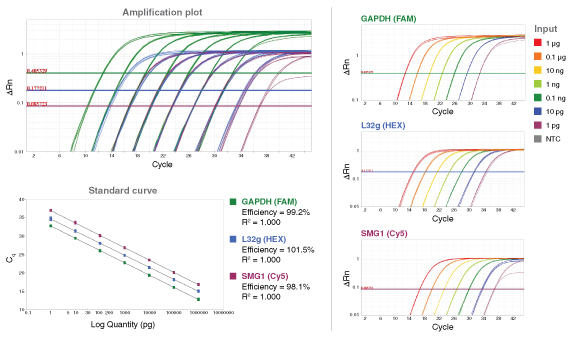
To detect or quantitate multiple targets in the same Luna reaction, select different fluorophores corresponding to separate detection channels of the real-time instrument. Include 400 nM of forward and reverse primer and 200 nM probe for each target to be detected in the reaction, and adjust concentrations if necessary based on performance (primer 200–900 nM, probe 100–500 nM). When loading qPCR protocol onto the real-time instrument, be sure to select the appropriate optical channels, as some instruments have a single channel recording mode that would prevent multiplex data collection and analysis. For ROX-dependent instruments, avoid ROX-labeled probes. The functionality of the primer and probe sets should be tested individually before attempting a multiplex reaction. When determining which fluorophores to include in a multiplex reaction, be sure to choose compatible reporter dyes and quenchers that have well separated fluorescence spectra or exhibit minimal overlap. - Amplicon Length
To ensure successful and consistent qPCR results, it is important to maximize PCR efficiency. An important aspect of this is the design of short PCR amplicons (typically 70–200 bp). Some optimization may be required (including the use of longer extension times), for targets that exceed that range. - Template Preparation and Concentration
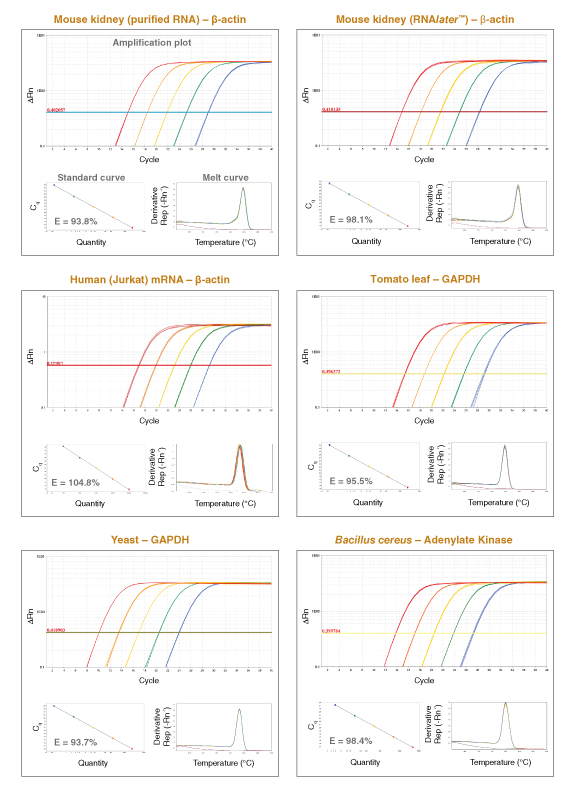
Luna qPCR is compatible with DNA samples prepared through typical nucleic acid purification methods. Prepared DNA should be stored in an EDTA-containing buffer (e.g., 1X TE) for long-term stability, and dilutions should be freshly prepared for a qPCR experiment by dilution into either TE or water.
Generally, a useful concentration of standard and unknown material will be in the range of 106 copies to 1 copy. For gDNA samples from large genomes (e.g., human, mouse) a range of 50 ng–1 pg of gDNA is typical. For small genomes, adjust as necessary using 106 –1 copy input as an approximate range. Note that for single copy dilutions, some samples will contain multiple copies and some will have none, as defined by the Poisson distribution.
For cDNA, use the product of a reaction containing 1 μg–0.1 pg starting RNA. cDNA does not need to be purified before addition to the Luna reaction but should be diluted at least 1:10 into the qPCR. - ROX Reference Dye
Some real-time instruments recommend the use of a passive reference dye (typically ROX) to overcome well-to-well variations that could be caused by machine limitations such as “edge effect”, bubbles, small differences in volume, and autofluorescence from dust or particulates in the reaction. However, ROX normalization does little to the variations caused by pipetting errors of templates/primers, heterogeneous mixing, and evaporation/condensation issues.
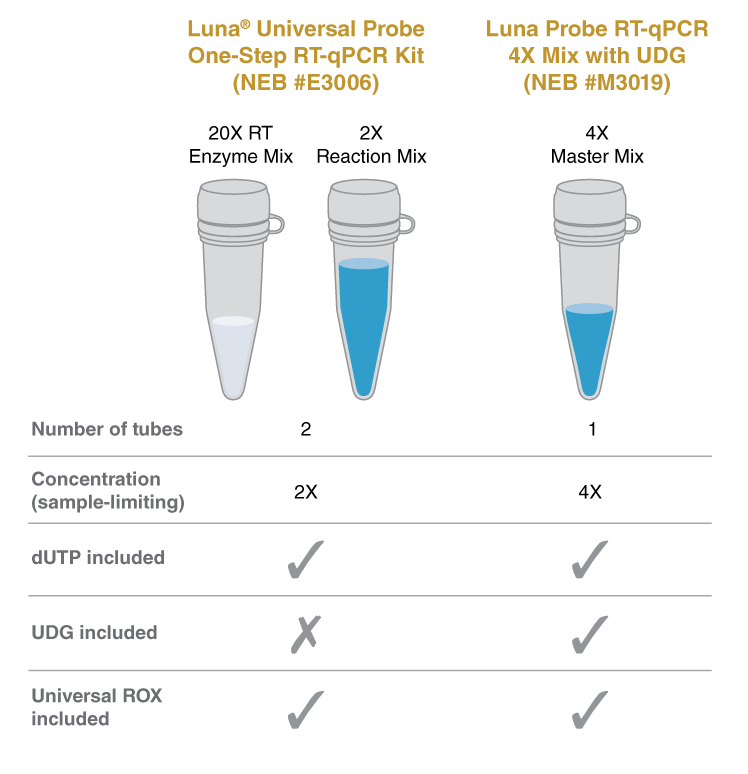
A universal passive reference dye is included in the following Luna® qPCR products: Luna Universal qPCR Master Mix (NEB #M3003), Luna Universal Probe qPCR Master Mix (NEB #M3004), Luna Universal One-Step RT-qPCR Kit (NEB #E3005), and Luna Universal Probe One-Step RT-qPCR Kit (NEB #E3006). These products support broad instrument compatibility (High-ROX, Low-ROX, ROX-independent) so no additional ROX is required for normalization.
The Luna Probe One-Step RT-qPCR Kit (No ROX) (E3007) contains no reference dye and is compatible with any instrument that does not require ROX. If ROX normalization is needed, ROX can be added. Please refer to instrument manufacturer’s instructions for greater details.
- Carryover Contamination Prevention
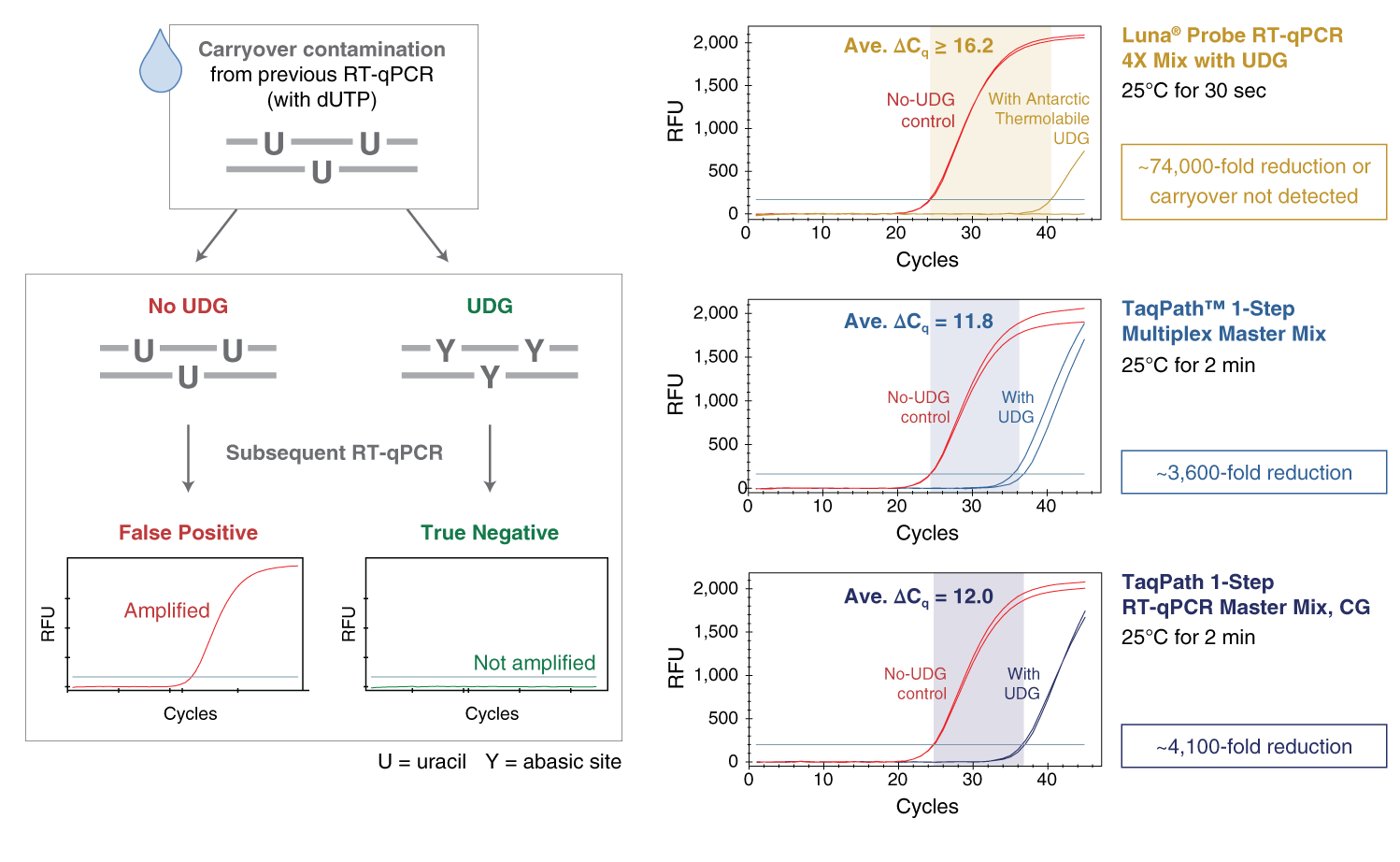
qPCR is an extremely sensitive method, and contamination in new qPCR assays with products from previous amplification reactions can cause a variety of issues such as false positive results and a decrease in sensitivity. The best way to prevent this “carryover” contamination is to practice good laboratory procedures and avoid opening the reaction vessel post amplification. However, to accommodate situations where additional anti-contamination measures are desired, the Luna Universal Probe qPCR Master Mix contains a mixture of dUTP/dTTP that results in the incorporation of dU into the DNA product during amplification. Pretreatment of qPCR experiments with uracil DNA glycosylase (UDG) will eliminate previously-amplified uracil-containing products by excising the uracil base to produce a non-amplifiable DNA product. The use of a thermolabile UDG is important, as complete inactivation of the UDG is required to prevent destruction of newly synthesized qPCR products.To enable carryover prevention, 0.025 units/μl Antarctic Thermolabile UDG (NEB #M0372) should be added to the reaction mix. To maximize elimination of contaminating products, set up the qPCR experiments at room temperature or include a 10 minute incubation step at 25°C before the initial denaturation step.
- Reaction Setup and Cycling Conditions
Due to the hot start nature of the polymerase, it is not necessary to preheat the thermocycler prior to use or set up reactions on ice.
For 96-well plates, we recommend a final reaction volume of 20 μl.
For 384-well plates, a final reaction volume of 10 μl is recommended.
When programming instrument cycling conditions, ensure a plate read is included at the end of the extension step, and a denaturation (melt) curve after cycling is complete to analyze product specificity.
Amplification for 40 cycles is sufficient for most applications, but for very low input samples 45 cycles may be used.
- Assay Design
操作说明、说明书 & 用法
-
操作说明
- Luna® Universal Probe qPCR Master Mix Protocol (M3004)
- Protocol for Two-step RT-qPCR using the LunaScript® RT SuperMix Kit (NEB #E3010) and the Luna® Universal qPCR Master Mix (NEB #M3003) or Luna Universal Probe qPCR Master Mix (NEB #M3004)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualM3004
-
使用指南
- Optimization Tips for Luna® qPCR
-
应用实例
- Probe-based qPCR Probe Compatibility and Multiplexing with Luna Universal Probe qPCR Master Mix
- High-throughput qPCR and RT-qPCR Workflows Enabled by Beckman Coulter Echo Acoustic Liquid Handling and NEB Luna Reagents
- Multiplex real-time PCR detection of monkeypox virus using Luna® qPCR Reagents
FAQs & 问题解决指南
-
FAQs
- How do I use qPCR to determine the concentration of my material?
- Can I set up my Luna® qPCR at room temperature?
- What is the difference between probe- and dye-based versions of the Luna® qPCR Mixes?
- Should I use probe- or dye-based detection for my qPCR assays?
- How should I design primers for Luna® qPCR?
- How long should my amplicon be for qPCR?
- Why is the Luna® qPCR Mix blue? Will this dye interfere with detection?
- Can I run the Luna® qPCR Mix on my qPCR instrument?
- Can I use fast instrument settings with the Luna® qPCR Mix?
- Do I need to add ROX?
- How many dilutions should I use to make a standard curve?
- Why does NEB recommend 40-45 cycles?
- Does the Luna® qPCR Mix contain dUTP? Can I use carryover contamination prevention methods?
- Can I run multiplex reactions with the Luna® Universal Probe Master Mix? Do I need to change my reaction conditions?
- Can I use a ROX-labeled probe with the Luna® Probe Mixes that contain a universal ROX reference dye?
- Can alternative probe based detection strategies be used with the Luna® Probe Mix?
- What samples can be used in qPCR with the Luna® Mix?
- Can I use cDNA? Does it matter how I make it?
- How much template material can I use in Luna® qPCR?
- How much primer and probe should I use with the Luna® Universal Probe qPCR Master Mix?
- Can I use shorter cycling times?
-
问题解决指南
- Luna® qPCR Troubleshooting Guide