上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息

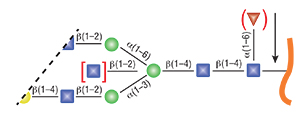
Peptide: N-Glycosidase F, also known as PNGase F, is a recombinant amidase which cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins (1).
产品来源
Cloned from Elizabethkingia miricola (formerly Flavobacterium meningosepticum) and expressed in E. coli (2).
特异性
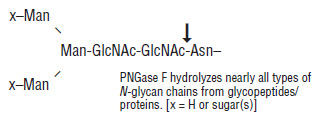
- 产品类别:
- Endoglycosidases Products,
- Proteome Analysis Products
- 应用:
- Expression Systems,
- Glycan Sequencing,
- Proteomics,
- Recombinant Glycoprotein Expression,
Glycoprotein Analysis
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
P0708S -20 PNGase F, Recombinant P0708SVIAL -20 1 x 0.03 ml 500,000 units/ml GlycoBuffer 2 B3704SVIAL -20 1 x 1 ml 10 X Glycoprotein Denaturing Buffer B1704SVIAL -20 1 x 1 ml 10 X NP-40 B2704SVIAL -20 1 x 1 ml 10 %
-
P0708L -20 PNGase F, Recombinant P0708LVIAL -20 1 x 0.15 ml 500,000 units/ml GlycoBuffer 2 B3704SVIAL -20 1 x 1 ml 10 X Glycoprotein Denaturing Buffer B1704SVIAL -20 1 x 1 ml 10 X NP-40 B2704SVIAL -20 1 x 1 ml 10 %
-
-
特性和用法
单位定义
One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 10 μg of denatured RNase B in 1 hour at 37°C in a total reaction volume of 10 μl.
Unit Definition Assay:
10 µg of RNase B are denatured with 1X Glycoprotein Denaturing Buffer (0.5% SDS, 40 mM DTT) at 100°C for 10 minutes. After the addition of NP-40 and GlycoBuffer 2, two-fold dilutions of PNGase F are added and the reaction mix is incubated for 1 hour at 37°C. Separation of reaction products are visualized by SDS-PAGE.1X Glycoprotein Denaturing Buffer
0.5% SDS
40 mM DTT1X NP-40
1% NP-40 in MilliQ-H2O反应条件
1X GlycoBuffer 2
Incubate at 37°C1X GlycoBuffer 2
50 mM Sodium Phosphate
(pH 7.5 @ 25°C)贮存溶液
50 mM NaCl
20 mM Tris-HCl
5 mM EDTA
50% Glycerol
pH 7.5 @ 25°C热失活
75°C for 10 minutes
分子量
实际: 36000 daltons
-
优势和特性
应用特性
- Removal of carbohydrate residues from proteins
-
相关产品
相关产品
- RNase B(对照底物)
- 糖苷内切酶反应缓冲液套装
- p0702-endo-h
- p0703-endo-hf
- p0706-remove-it-pngase-f
- p0733-o-glycosidase
-
注意事项
- To deglycosylate a native glycoprotein, longer incubation time as well as more enzyme may be required.
- Since PNGase F, Recombinant activity is inhibited by SDS, it is essential to have NP-40 in the reaction mixture. It is not known why this non-ionic detergent counteracts the SDS inhibition at the present time.
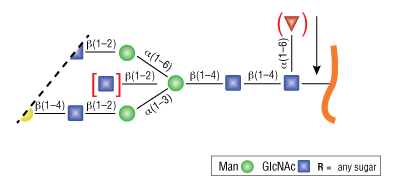
- PNGase F, Recombinant will not cleave N-linked glycans containing core α1-3 Fucose.
-
参考文献
- Maley, F. et al. (1989). Anal. Biochem. 180, 195-204.
- Chen, M. New England Biolabs, Inc., Unpublished observation
操作说明、说明书 & 用法
-
操作说明
- PNGase F Protocol
-
使用指南
- Detailed Characterization of Several Glycosidase Enzymes
- Glycobiology Unit Conversion Chart
-
应用实例
- AppNote_Remove-iT_PNGase_F_Effective_Release_and_Recovery_of_Neutral_and_Sialylated_N-glycans
- AppNote_Unbiased_and_Fast_IgG_Deglycosylation _for_Accurate_N-glycan_Analysis_using_Rapid_PNGase_F
- AppNote_Proteomics_Fast_and_Efficient_Antibody_Deglycosylation_using_Rapid_PNGase_F
- AppNote_Glycan_Analysis_of_Murine_IgG2a_by_Enzymatic_Digestion_with_PNGase_F_and_Trypsin
- Glycan Analysis of Murine IgG2a by Enzymatic Digestion with PNGase F and Trypsin, Followed by Mass Spectrometric Analysis
- Detailed Characterization of Antibody Glycan Structure using the N-Glycan Sequencing Kit
- Characterization of Glycans from Erbitux®, Rituxan® and Enbrel® using PNGase F (Glycerol-free), Recombinant
- Remove iT PNGase F Effective Release and Recovery of Neutral and Sialylated N glycans
工具 & 资源
-
选择指南
- Endoglycosidase Selection Chart
FAQs & 问题解决指南
-
FAQs
- Is there a difference between PNGase F (P0704/P0705) and PNGase F, Recombinant (P0708/P0709)?
- I tried deglycosylating my glycoprotein with Remove-iT® PNGase F but did not see removal of the carbohydrate. What could be the problem?
- What happens to the asparagine after PNGase removes the sugar?
- Why is protein degraded? When I denature and add SDS all I see on my SDS-PAGE is a smear or no protein. Can a protease inhibitor cocktail be used in a PNGase F, Recombinant reaction?
- What is the difference between PNGase F, Endo H and O-Glycosidase?
- Does PNGase F work in Urea?
- What are the typical reaction conditions for PNGase F, Recombinant?
- How much PNGase F, Recombinant should I use to remove my carbohydrate under native or DTT denaturing conditions?
- How do I inhibit PNGase F, Recombinant?
- Is PNGase F, Recombinant compatible with downstream analysis such as HPLC and Mass Spectrometry?
- What are Glycosidases and their uses?
- Do detergents inhibit exoglycosidases/endoglycosidases?
- Why have the NEB Glycosidase enzymes changed reaction buffers? What are the new reaction buffers and can I still use an enzyme with its old buffer? Where can I find the composition of the old buffers?
- What is a good endoglycosidase substrate?
-
实验技巧
- You can use this enzyme under native or denaturing conditions
- Under native conditions we recommend adding more enzyme and using longer incubation times
- PNGase F activity is inhibited by SDS, therefore under denaturing conditions it is essential to have NP-40 present in the reaction mixture in a 1:1 ratio.
- PNGase F will not cleave N-linked glycans containing core α1-3 Fucose (PNGase A must be used in this instance)
- A good positive control substrate is RNase B