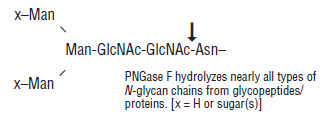上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
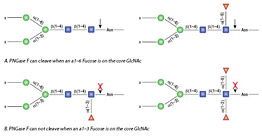
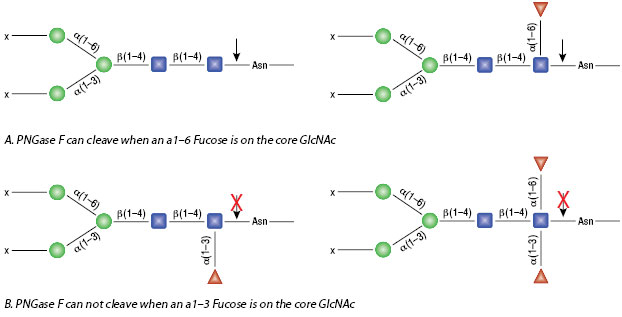
PNGase A is a recombinant amidase, which cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and short complex oligosaccharides such as those found in plant and insect cells from N-linked glycoproteins and glycopeptides. PNGase A differs from PNGase F in that it cleaves N-linked glycans with or without α(1,3)-linked core fucose residues.

产品来源
Cloned from Oryza sativa (rice) and expressed in Pichia pastoris.
- 产品类别:
- Endoglycosidases Products,
- Proteome Analysis Products
- 应用:
- Glycomics and glycoproteomics,
- Expression Systems,
- Glycan Sequencing,
- Proteomics,
- Recombinant Glycoprotein Expression,
Glycoprotein Analysis
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
P0707S 4 PNGase A P0707SVIAL 4 1 x 0.03 ml 5,000 units/ml GlycoBuffer 3 B1720SVIAL -20 1 x 1 ml 10 X Glycoprotein Denaturing Buffer B1704SVIAL -20 1 x 1 ml 10 X NP-40 B2704SVIAL -20 1 x 1 ml 10 %
-
P0707L 4 PNGase A P0707LVIAL 4 1 x 0.15 ml 5,000 units/ml GlycoBuffer 3 B1720SVIAL -20 1 x 1 ml 10 X Glycoprotein Denaturing Buffer B1704SVIAL -20 1 x 1 ml 10 X NP-40 B2704SVIAL -20 1 x 1 ml 10 %
-
-
特性和用法
单位定义
One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 1 µg of denatured recombinant Avidin produced in Maize in 1 hour at 37°C in a total reaction volume of 10 µl.
反应条件
1X GlycoBuffer 3
Incubate at 37°C1X GlycoBuffer 3
50 mM sodium acetate
(pH 6 @ 25°C)贮存溶液
20 mM Tris-HCl
50 mM NaCl
5 mM EDTA
pH 7.5 @ 25°C热失活
65°C for 10 minutes
分子量
实际: 63.8 kDa
单位活性检测条件
1 μg of recombinant Avidin is denatured with 1X Glycoprotein Denaturing Buffer at 100°C for 10 minutes. After the addition of NP-40 and GlycoBuffer 3, two-fold dilutions of PNGase A are added and the reaction mix is incubated for 1 hour at 37°C. Separation of reaction products are visualized by SDS-PAGE.
-
相关产品
相关产品
- p0733-o-glycosidase
- 蛋白去糖基化混合液 II
- p0705-pngase-f-glycerol-free
- Rapid 快速 PNGase F
- p0711-rapid-pngase-f-non-reducing-format
-
注意事项
- PNGase A is active on both glycoproteins and glycopeptides.
- PNGase A cannot cleave larger N-glycans such as those from Fetuin, Fibrinogen, IgG, Lactoferrin and Transferrin.
- PNGase A is able to cleave high mannose N-glycan structures from Man 3 up to Man 9.
- Reactions may be scaled-up linearly to accommodate larger reaction volumes.
操作说明、说明书 & 用法
-
操作说明
- Reaction Conditions for PNGase A (P0707)
工具 & 资源
-
选择指南
- Endoglycosidase Selection Chart
FAQs & 问题解决指南
-
FAQs
- What is the difference between PNGase F and PNGase A?
- Can PNGase A be used under non-denaturing (native) conditions?
- Which high mannose structures can PNGase A cleave?
- Can PNGase A cleave large, complex oligosaccharides?
- What happens to the asparagine after PNGase A removes the sugar?
- What is a good PNGase A substrate?
- Is PNGase A compatible with downstream analysis such as HPLC and Mass Spectrometry?
- How much PNGase A should I use to remove my carbohydrate under native or DTT denaturing conditions?
- Why is my protein degraded? When I denature and add SDS all I see on my SDS-PAGE is a smear or no protein. Can a protease inhibitor cocktail be used in a PNGase A reaction?
- What are Glycosidases and their uses?
-
实验技巧
NEB’s version of PNGase A can cleave glycoproteins, there is no need for tryptic digest prior to deglycosylation.
PNGase A can cleave N-linked glycans containing core α1-3 Fucose.
PNGase A activity is inhibited by SDS, therefore under denaturing conditions it is essential to have NP-40 present in the reaction mixture in a 1:1 ratio.
A good positive control substrate is recombinant avidin from maize or HRP.