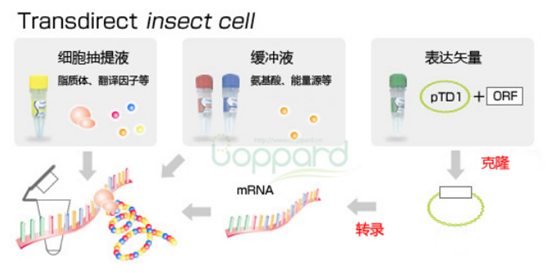
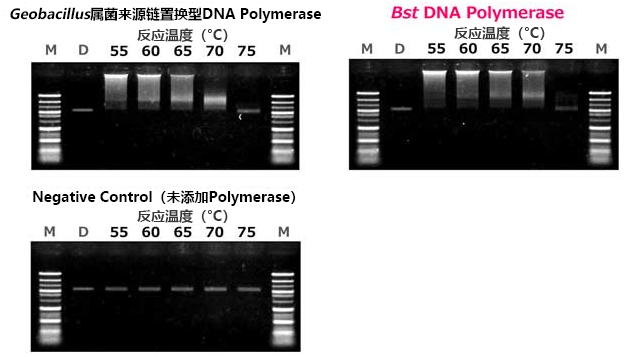
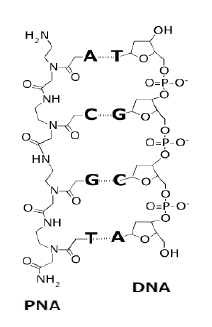
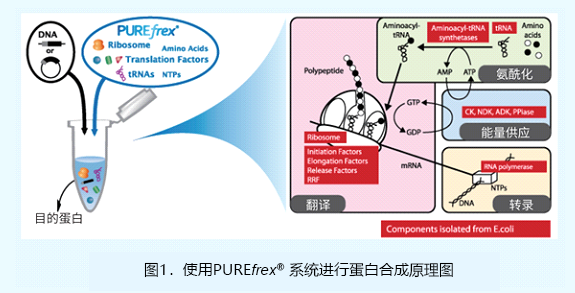
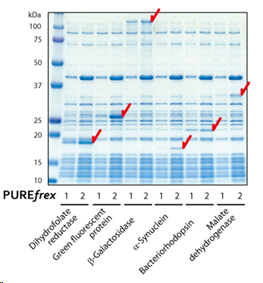
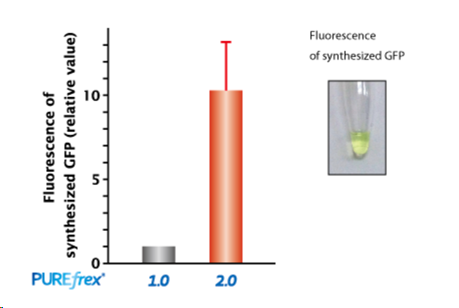
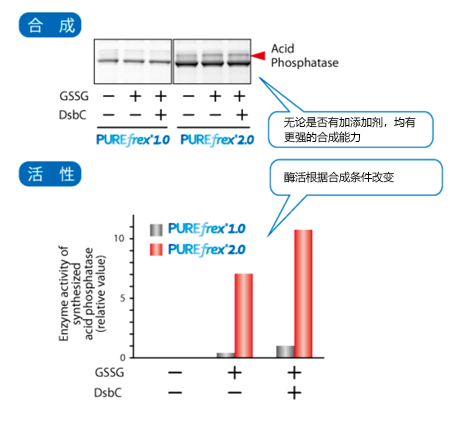
| [1] |
Murakami, S., Matsumoto, R., & Kanamori, T.. (2019). Constructive approach for synthesis of a functional IgG using a reconstituted cell-free protein synthesis system. Scientific reports 9(1), 671. |
| [2] |
Doerr, A., de Reus, E., van Nies, P., van der Haar, M., Wei, K., Kattan, J., et al. (2019). Modelling cell-free RNA and protein synthesis with minimal systems. Physical biology, 16, 025001. |
| [3] |
Dopp, J., Tamiev, D., & Reuel, N. F.. (2019). Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnology advances, 37(1), 246-258. |
| [4] |
Marsden, A. P., Hollins, J. J., O’Neill, C., Ryzhov, P., Higson, S., Mendonça, C. A., et al. (2018). Investigating the Effect of Chain Connectivity on the Folding of a Beta-Sheet Protein On and Off the Ribosome. Journal of molecular biology, 430, 5207-5216. |
| [5] |
Tian, P., Steward, A., Kudva, R., Su, T., Shilling, P. J., Nickson, A. A., et al. (2018). The Folding Pathway of an Ig Domain is Conserved On and Off the Ribosome. Proceedings of the National Academy of Sciences, 201810523., 115(48), E11284-E11293. |
| [6] |
Gessesse, B., Nagaike, T., Nagata, K., Shimizu, Y., & Ueda, T.. (2018). G-Protein Coupled Receptor Protein Synthesis on a Lipid Bilayer Using a Reconstituted Cell-Free Protein Synthesis System. Life, 8(4), 54. |
| [7] |
Kamiya, N., Ohama, Y., Minamihata, K., Wakabayashi, R., & Goto, M.. (2018). Liquid Marbles as an Easy‐to‐Handle Compartment for Cell‐Free Synthesis and In Situ Immobilization of Recombinant Proteins. Biotechnology journal,13(12). |
| [8] |
Hayase, G., & Nomura, S. I. M.. (2018). Large-Scale Preparation of Giant Vesicles by Squeezing a Lipid-Coated Marshmallow-like Silicone Gel in a Buffer. Langmuir, 34(37), 11021-11026. |
| [9] |
Fujiwara, K., Ito, K., & Chiba, S.. (2018). MifM-instructed translation arrest involves nascent chain interactions with the exterior as well as the interior of the ribosome. Scientific reports, 8(1), 10311. |
| [10] |
Sugimoto, S., Arita-Morioka, K. I., Terao, A., Yamanaka, K., Ogura, T., & Mizunoe, Y.. (2018). Multitasking of Hsp70 chaperone in the biogenesis of bacterial functional amyloids. Communications Biology, 1(1), 52. |
| [11] |
Kamiya, Y., Arimura, Y., Ooi, H., Kato, K., Liang, X. G., & Asanuma, H.. (2018). Development of Visible‐Light‐Responsive RNA Scissors Based on a 10–23 DNAzyme. ChemBioChem. 19, 1305-1311. |
| [12] |
Fujii, S., Sawa, T., Motohashi, H., & Akaike, T.. (2018). Persulfide synthases that are functionally coupled with translation mediate sulfur respiration in mammalian cells. British Journal of Pharmacology, 176(4), 607-615. |
| [13] |
Komura, R., Aoki, W., Motone, K., Satomura, A., & Ueda, M.. (2018). High-throughput evaluation of T7 promoter variants using biased randomization and DNA barcoding. PLOS ONE, 13(5), e0196905. |
| [14] |
van Nies, P., Westerlaken, I., Blanken, D., Salas, M., Mencía, M., & Danelon, C.. (2018). Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nature communications, 9(1), 1583. |
| [15] |
Furusato, T., Horie, F., Matsubayashi, H. T., Amikura, K., Kuruma, Y., & Ueda, T.. (2018). De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS synthetic biology, 7(4), 953-961. |
| [16] |
Natan, E., Endoh, T., Haim-Vilmovsky, L., Flock, T., Chalancon, G., Hopper, J. T., et al. (2018). Cotranslational protein assembly imposes evolutionary constraints on homomeric proteins. Nature structural & molecular biology, 25(3), 279. |
| [17] |
Ito, N., Katoh, K., Kushige, H., Saito, Y., Umemoto, T., Matsuzaki, Y., et al. (2018). Ribosome incorporation into somatic cells promotes lineage transdifferentiation towards multipotency. Scientific reports, 8(1), 1634. |
| [18] |
Reyes, S. G., Kuruma, Y., & Tsuda, S.. (2017). Uncovering cell-free protein expression dynamics by a promoter library with diverse strengths. bioRxiv, 214593. |
| [19] |
Katano, Y., Li, T., Baba, M., Nakamura, M., Ito, M., Kojima, K., et al. (2017). Generation of thermostable Moloney murine leukemia virus reverse transcriptase variants using site saturation mutagenesis library and cell-free protein expression system. Bioscience, biotechnology, and biochemistry, 81(12), 2339-2345. |
| [20] |
Chadani, Y., Niwa, T., Izumi, T., Sugata, N., Nagao, A., Suzuki, T., et al. (2017). Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing. Molecular cell, 68(3), 528-539. |
| [21] |
Akaike, T., Ida, T., Wei, F. Y., Nishida, M., Kumagai, Y., Alam, M. M., et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nature communications, 8(1), 1177. |
| [22] |
Shepherd, T. R., Du, L., Liljeruhm, J., Wang, J., Sjödin, M. O., Wetterhall, M., et al. (2017). De novo design and synthesis of a 30-cistron translation-factor module. Nucleic acids research, 45(18), 10895-10905. |
| [23] |
Matsumoto, K. I., Yamazaki, K., Kawakami, S., Miyoshi, D., Ooi, T., Hashimoto, S., & Taguchi, S.. (2017). In vivo target exploration of apidaecin based on Acquired Resistance induced by Gene Overexpression (ARGO assay). Scientific reports, 7(1), 12136. |
| [24] |
Judd, J., Boucher, N., Van Roey, E., Gray, T. A., & Derbyshire, K. M.. (2017). Application of distributive conjugal DNA transfer in Mycobacterium smegmatis to establish a genome-wide synthetic genetic array. Journal of Bacteriology, 199(20). |
| [25] |
Goto, Y., Murakami, H., & Suga, H.. (2008). Initiating translation with D-amino acids. RNA, 14(7), 1390–1398. |
| [26] |
Ueta, M., Wada, C., Bessho, Y., Maeda, M., & Wada, A.. (2017). Ribosomal protein L31 in Escherichia coli contributes to ribosome subunit association and translation, whereas short L31 cleaved by protease 7 reduces both activities. Genes to Cells, 22(5), 452-471. |
| [27] |
Nilsson, O. B., Nickson, A. A., Hollins, J. J., Wickles, S., Steward, A., Beckmann, R., et al. (2017). Cotranslational folding of spectrin domains via partially structured states. Nature structural & molecular biology, 24(3), 221. |
| [28] |
Fan, Y., Hoshino, T., & Nakamura, A.. (2017). Identification of a VapBC toxin–antitoxin system in a thermophilic bacterium Thermus thermophilus HB27. Extremophiles, 21(1), 153-161. |
| [29] |
Scott, A., Noga, M. J., de Graaf, P., Westerlaken, I., Yildirim, E., & Danelon, C.. (2016). Cell-free phospholipid biosynthesis by gene-encoded enzymes reconstituted in liposomes. PloS one, 11(10), e0163058. |
| [30] |
Nakayama, M., Komiya, S., Fujiwara, K., Horisawa, K., & Doi, N.. (2016). In vitro selection of bispecific diabody fragments using covalent bicistronic DNA display. Biochemical and biophysical research communications, 478(2), 606-611. |
| [31] |
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., & Ueda, T.. (2001). Cell-free translation reconstituted with purified components. Nature Biotechnology, 19(8), 751-755. |
| [32] |
Radomska, K. A., Ordoñez, S. R., Wösten, M. M., Wagenaar, J. A., & van Putten, J. P.. (2016). Feedback control of Campylobacter jejuni flagellin levels through reciprocal binding of FliW to flagellin and the global regulator CsrA. Molecular microbiology, 102(2), 207-220. |
| [33] |
Nilsson, O. B., Müllerlucks, A., Kramer, G., Bukau, B., & Heijne, G. V.. (2016). Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. Journal of Molecular Biology, 428(6), 1356-1364. |
| [34] |
Chadani, Y., Niwa, T., Chiba, S., Taguchi, H., & Ito, K.. (2016). Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing. Proceedings of the National Academy of Sciences, 113(7), E829–E838. |
| [35] |
Ando, M., Akiyama, M., Okuno, D., Hirano, M., Ide, T., Sawada, S., et al. (2016). Liposome chaperon in cell-free membrane protein synthesis: one-step preparation of KcsA-integrated liposomes and electrophysiological analysis by the planar bilayer method. Biomaterials science, 4(2), 258-264. |
| [36] |
Shiraishi, A., Mochizuki, S., Miyakoshi, A., Kojoh, K., & Okada, Y.. (2016). Development of human neutralizing antibody to ADAMTS4 (aggrecanase-1) and ADAMTS5 (aggrecanase-2). Biochemical and biophysical research communications, 469(1), 62-69. |
| [37] |
Nagumo, Y., Fujiwara, K., Horisawa, K., Yanagawa, H., & Doi, N.. (2015). PURE mRNA display for in vitro selection of single-chain antibodies. The Journal of Biochemistry, 159(5), 519-526. |
| [38] |
Niwa, T., Sasaki, Y., Uemura, E., Nakamura, S., Akiyama, M., Ando, M.,et al. (2015). Comprehensive study of liposome-assisted synthesis of membrane proteins using a reconstituted cell-free translation system. Scientific reports, 5(1), 18025. |
| [39] |
Yamamoto, H., Shima, T., Yamaguchi, M., Mochizuki, Y., Hoshida, H., Kakuta, S.,et al. (2015). The thermotolerant yeast Kluyveromyces marxianus is a useful organism for structural and biochemical studies of autophagy. Journal of Biological Chemistry, 290(49), 29506–29518. |
| [40] |
Ishii, E., Chiba, S., Hashimoto, N., Kojima, S., Homma, M., Ito, K., et al. (2015). Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria.Proceedings of the National Academy of Sciences, 112(40), E5513-E5522. |
| [41] |
Nilsson, O. B., Hedman, R., Marino, J., Wickles, S., Bischoff, L., Johansson, M., et al. (2015). Cotranslational protein folding inside the ribosome exit tunnel. Cell reports, 12(10), 1533-1540. |
| [42] |
Kuruma, Y., & Ueda, T.. (2016). Corrigendum: the pure system for the cell-free synthesis of membrane proteins. Nature Protocols, 11(3), 616. |
| [43] |
Morita, M., Onoe, H., Yanagisawa, M., Ito, H., Ichikawa, M., Fujiwara, K., et al. (2015). Droplet‐Shooting and Size‐Filtration (DSSF) Method for Synthesis of Cell‐Sized Liposomes with Controlled Lipid Compositions. ChemBioChem, 16(14), 2029-2035. |
| [44] |
Yamashita, H., Morita, M., Sugiura, H., Fujiwara, K., Onoe, H., & Takinoue, M.. (2015). Generation of monodisperse cell-sized microdroplets using a centrifuge-based axisymmetric co-flowing microfluidic device. Journal of bioscience and bioengineering, 119(4), 492-495. |
| [45] |
Nies, V., & Pauline.. (2015). monitoring mrna and protein levels in bulk and in model vesicle-based artificial cells. Methods in Enzymology, 550, 187-214. |
| [46] |
Ichihashi, N., Kobori, S., & Yomo, T..(2015). Simple Identification of Two Causes of Noise in an Aptazyme System by Monitoring Cell-Free Transcription. Methods in Enzymology, 550, 93-107. |
| [47] |
Kogure, H., Handa, Y., Nagata, M., Kanai, N., Peter Güntert, & Kubota, K., et al. (2014). Identification of residues required for stalled-ribosome rescue in the codon-independent release factor yaej. Nucleic Acids Research, 42(5), 3152. |
| [48] |
Shimizu, Y., Kuruma, Y., Kanamori, T., & Ueda, T.. (2014). The pure system for protein production. Methods in Molecular Biology, 1118(1118), 275-284. |
| [49] |
Jackson, K., Kanamori, T., Ueda, T., & Fan, Z. H.. (2014). Protein synthesis yield increased 72 times in the cell-free pure system. Integrative Biology, 6(8),781-788. |
| [50] |
Matsubayashi, H., Kuruma, Y., & Ueda, T.. (2014). In vitro synthesis of the e. coli sec translocon from dna. Angewandte Chemie International Edition in English, 53(29), 7535-7538. |
| [51] |
Nourian, Z., Scott, A., & Danelon, C.. (2014). Toward the assembly of a minimal divisome. Systems and Synthetic Biology, 8(3), 237-247. |
| [52] |
Sugimoto, N.. (2014). Noncanonical structures and their thermodynamics of dna and rna under molecular crowding: beyond the watson-crick double helix. Int Rev Cell Mol Biol, 307, 205-273. |
| [53] |
Fujiwara, K., Katayama, T., & Nomura, S. I.. (2013). Cooperative working of bacterial chromosome replication proteins generated by a reconstituted protein expression system. Nucleic Acids Research, 41(14), 7176-7183. |
| [54] |
Endoh, T., Kawasaki, Y., & Sugimoto, N.. (2013). Translational halt during elongation caused by g-quadruplex formed by mrna. Methods, 64(1), 73-78. |
| [55] |
Hong, S. H., Ntai, I., Haimovich, A. D., Kelleher, N. L., Isaacs, F. J., & Jewett, M. C.. (2014). Cell-free protein synthesis from a release factor 1 deficient, escherichia coli, activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synthetic Biology, 3(6), 398-409. |
| [56] |
Chizzolini, F., Forlin, M., Cecchi, D., & Mansy, S. S.. (2013). Gene position more strongly influences cell-free protein expression from operons than t7 transcriptional promoter strength. ACS Synthetic Biology, 3(6). |
| [57] |
Fujii, S., Matsuura, T., Sunami, T., Kazuta, Y., & Yomo, T.. (2013). In vitro evolution of -hemolysin using a liposome display. Proceedings of the National Academy of Sciences, 110(42), 16796-16801. |
| [58] |
Nies, V., Pauline, Nourian, Zohreh, Kok, & Maurits, et al. (2013). Unbiased tracking of the progression of mrna and protein synthesis in; bulk and in liposome-confined reactions. Chembiochem A European Journal of Chemical Biology, 14(15), 1963-1966. |
| [59] |
Niederholtmeyer, H., Stepanova, V., & Maerkl, S. J.. (2013). Implementation of cell-free biological networks at steady state. Proceedings of the National Academy of Sciences, 110(40), 15985-15990. |
| [60] |
Lentini, R., Forlin, M., Martini, L., Bianco, C. D., Spencer, A. C., & Torino, D., et al. (2013). Fluorescent proteins and in vitro genetic organization for cell-free synthetic biology. ACS Synthetic Biology, 2(9), 482-489. |
| [61] |
Woolstenhulme, C. J., Parajuli, S., Healey, D. W., Valverde, D. P., Petersen, E. N., & Starosta, A. L., et al. (2013). Nascent peptides that block protein synthesis in bacteria. Proceedings of the National Academy of Sciences, 110(10), E878-E887. |
| [62] |
Jewett, M. C., Fritz, B. R., Timmerman, L. E., & Church, G. M.. (2014). In vitro integration of ribosomal rna synthesis, ribosome assembly, and translation. Molecular Systems Biology, 9(1), 678-678. |
| [63] |
Niederholtmeyer, H., Xu, L., & Maerkl, S. J.. (2013). Real-time mrna measurementduring an in vitro transcription and translationreaction using binary probes. ACS Synthetic Biology, 2(8), 411-417. |
| [64] |
Endoh, T., Kawasaki, Y., & Sugimoto, N.. (2013). Stability of rna quadruplex in open reading frame determines proteolysis of human estrogen receptor α. Nucleic Acids Research, 41(12), 6222-6231. |
| [65] |
Endoh, T., Kawasaki, Y., & Sugimoto, N.. (2013). Suppression of gene expression by g-quadruplexes in open reading frames depends on g-quadruplex stability. Angewandte Chemie International Edition, 52(21), 5522-5526. |
| [66] |
Lee, K. B., Kim, H. C., Kim, D. M., Kang, T. J., & Suga, H.. (2013). Comparative evaluation of two cell-free protein synthesis systems derived from escherichia coli for genetic code reprogramming. Journal of Biotechnology, 164(2), 330-335. |
| [67] |
Nakamura, Y., Ogura, M., Ogura, K., Tanaka, D., & Inagaki, N.. (2012). Sirt5 deacetylates and activates urate oxidase in liver mitochondria of mice. FEBS letters, 586(23), 4076-4081. |
| [68] |
Fujino, Y., Fujita, R., Wada, K., Fujishige, K., & Ueda, T.. (2012). Robust in vitro affinity maturation strategy based on interface-focused high-throughput mutational scanning. Biochemical and Biophysical Research Communications, 428(3), 395-400. |
| [69] |
Venancio-Marques, A., Liu, Y.-J., Diguet, A., di Maio, T., Gautier, A., & Baigl, D. (2012). Modification-Free Photocontrol of β-Lactam Conversion with Spatiotemporal Resolution. ACS Synthetic Biology, 1(11), 526–531. |
| [70] |
Nicolini, C., Bragazzi, N., & Pechkova, E.. (2012). Nanoproteomics enabling personalized nanomedicine. Advanced Drug Delivery Reviews, 64(13), 1522-1531. |
| [71] |
Matsuura, T., Hosoda, K., Kazuta, Y., Ichihashi, N., Suzuki, H., & Yomo, T.. (2012). Effects of compartment size on the kinetics of intracompartmental multimeric protein synthesis. ACS Synthetic Biology, 1(9), 431-437. |
| [72] |
Ong, H. J., Siau, J. W., Zhang, J. B., Hong, M., Flotow, H., & Ghadessy, F.. (2012). Analysis of p53 binding to dna by fluorescence imaging microscopy. Micron, 43(9), 996-1000. |
| [73] |
Shimizu, Y.. (2012). Arfa recruits rf2 into stalled ribosomes. Journal of molecular biology, 423(4), 624-631. |
| [74] |
Nagano, T., Kojima, K., Hisabori, T., Hayashi, H., Morita, E. H., & Kanamori, T., et al. (2012). Elongation factor g is a critical target during oxidative damage to the translation system of escherichia coli. Journal of Biological Chemistry, 287(34), 28697-28704. |
| [75] |
Ying, & B.-W. (2003). A novel screening system for self-mrna targeting proteins. Journal of Biochemistry, 133(4), 485-491. |
| [76] |
Kobori, S., Ichihashi, N., Kazuta, Y., Matsuura, T., & Yomo, T.. (2012). Kinetic analysis of aptazyme-regulated gene expression in a cell-free translation system: modeling of ligand-dependent and -independent expression. Rna-a Publication of the Rna Society, 18(8), 1458-1465. |
| [77] |
Bruder, J., Siewert, K., Obermeier, B., Malotka, J., Scheinert, P., & Kellermann, J., et al. (2012). Target specificity of an autoreactive pathogenic human γδ-T cell receptor in myositis. Journal of Biological Chemistry, 287(25), 20986-20995. |
| [78] |
Nishimura, K., Matsuura, T., Nishimura, K., Sunami, T., Suzuki, H., & Yomo, T.. (2012). Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir, 28(22), 8426-8432. |
| [79] |
Okano, T., Matsuura, T., Kazuta, Y., Suzuki, H., & Yomo, T.. (2012). Cell-free protein synthesis from a single copy of dna in a glass microchamber. Lab on a Chip, 12(15), 2704. |
| [80] |
Guarino, C., & Delisa, M. P.. (2012). A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology, 22(5), 596-601. |
| [81] |
Stögbauer, T., Windhager, L., Zimmer, R., & Rädler, J. O. (2012). Experiment and mathematical modeling of gene expression dynamics in a cell-free system. Integrative Biology, 4(5), 494-501. |
| [82] |
Do, P. M., Varanasi, L., Fan, S., Li, C., Kubacka, I., & Newman, V., et al. (2012). Mutant p53 cooperates with ets2 to promote etoposide resistance. Genes & Development, 26(8), 830-845. |
| [83] |
Kriechbaumer, V., Wang, P., Hawes, C., & Abell, B. M.. (2012). Alternative splicing of the auxin biosynthesis gene yucca4 determines its subcellular compartmentation. The Plant Journal, 70(2), 292-302. |
| [84] |
Zhu, X., Ahmad, S. M., Aboukhalil, A., Busser, B. W., & Michelson, A. M.. (2012). Differential regulation of mesodermal gene expression by drosophila cell type-specific forkhead transcription factors. Development, 139(8), 1457-1466. |
| [85] |
Guillen Schlippe, Y. V., Hartman, M. C. T., Josephson, K., & Szostak, J. W.. (2012). in vitror, selection of highly modified cyclic peptides that act as tight binding inhibitors. Journal of the American Chemical Society, 134(25), 10469-10477. |
| [86] |
Takahashi, S., Tsuji, K., Ueda, T., & Okahata, Y.. (2012). Traveling time of a translating ribosome along messenger rna monitored directly on a quartz crystal microbalance. Journal of the American Chemical Society, 134(15), 6793-6800. |
| [87] |
Papenfort, K., Podkaminski, D., Hinton, J. C. D., & Jörg Vogel. (2012). The ancestral sgrs rna discriminates horizontally acquired salmonella mrnas through a single g-u wobble pair. Proceedings of the National Academy of Sciences, 109(13), E757-764. |
| [88] |
Danelon, C., Nourian, Z., Roelofsen, W., & Westerlaken, I.. (2012). Triggered gene expression in fed-vesicle microreactors with a multifunctional membrane. Biophysical Journal, 102(3), 715a. |
| [89] |
Rosenblum, G., Chen, C., Kaur, J., Cui, X., Goldman, Y. E., & Cooperman, B. S.. (2012). Real-time assay for testing components of protein synthesis. Nucleic Acids Research, 40(12), e88-e88. |
| [90] |
Machida, K., Masutani, M., Kobayashi, T., Mikami, S., Nishino, Y., & Miyazawa, A., et al. (2012). Reconstitution of the human chaperonin cct by co-expression of the eight distinct subunits in mammalian cells. Protein Expression & Purification, 82(1), 61-69. |
| [91] |
Barendt, P. A., Shah, N. A., Barendt, G. A., Sarkar, C. A., & Hughes, D.. (2012). Broad-specificity mrna–rrna complementarity in efficient protein translation. PLoS Genetics, 8(3), e1002598. |
| [92] |
Wang, H. H., Huang, P.-Y., Xu, G., Haas, W., Marblestone, A., Li, J. et al.. (2012). Multiplexed in Vivo His-Tagging of Enzyme Pathways for in Vitro Single-Pot Multienzyme Catalysis. ACS Synthetic Biology, 1(2), 43–52. |
| [93] |
Holmqvist, E., Unoson, C., Reimegård, J., & Wagner, E. G. H. (2012). A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Molecular Microbiology, 84(3), 414–427. |
| [94] |
Endoh, T., Kawasaki, Y., & Sugimoto, N.. (2012). Synchronized translationfor detection of temporalstalling of ribosome during single-turnover translation. Analytical Chemistry, 84(2), 857-861. |
| [95] |
Marcin, D., Reynolds, C. B., & Fairweather, N. F.. (2012). Clostridium difficile cell wall protein cwpv undergoes enzyme-independent intramolecular autoproteolysis. Journal of Biological Chemistry, 287(2), 1538-1544. |
| [96] |
Atsushi, O., Masayoshi, H., Shinsuke, S., & Yasuhiro, A.. (2012). A concept for selection of codon-suppressor trnas based on read-through ribosome display in an in vitro compartmentalized cell-free translation system. Journal of Nucleic Acids, 2012, 538129. |
| [97] |
Lazzeriniospri, L., Stano, P., Luisi, P. L., & Marangoni, R.. (2012). Characterization of the emergent properties of a synthetic quasi-cellular system. Bmc Bioinformatics, 13(Suppl 4), S9. |
| [98] |
Nobuhide, D., Natsuko, Y., Hideaki, M., Yasutsugu, Y., Tetsuya, N., & Nobutaka, M., et al. (2012). Dna display selection of peptide ligands for a full-length human g protein-coupled receptor on cho-k1 cells. PLoS ONE, 7(1), e30084. |
| [99] |
Harada, R., Furumoto, S., Yoshikawa, T., Ishikawa, Y., Shibuya, K., & Okamura, N., et al. (2012). Synthesis of [11c]interleukin 8 using a cell-free translation system and l-[11c]methionine. Nuclear Medicine & Biology, 39(1), 155-160. |
| [100] |
Wang, X., Morgan, R., Nugent, M. L., Gupta, Y., Xu, S., & Fomenkov, A., et al. (2011). Characterization of type ii and iii restriction-modification systems from bacillus cereus strains atcc 10987 and atcc 14579. Journal of Bacteriology, 194(1), 49-60. |
| [101] |
Hufton, S. E.. (2012). Affinity maturation and functional dissection of a humanised anti-rage monoclonal antibody by ribosome display. Methods in Molecular Biology, 805, 403-422. |
| [102] |
Ohashi, H., Kanamori, T., Osada, E., Akbar, B. K., & Ueda, T.. (2012). Peptide screening using pure ribosome display. Methods in Molecular Biology, 805(1), 251-259. |
| [103] |
Nishikawa, T., Sunami, T., Matsuura, T., & Yomo, T. (2012). Directed Evolution of Proteins throughIn VitroProtein Synthesis in Liposomes. Journal of Nucleic Acids, 2012, 1–11. |
| [104] |
Takeshi, S., Hiroshi, Y., & Nobuhide, D.. (2012). in vitro selection of fab fragments by mrna display and gene-linking emulsion pcr. Journal of Nucleic Acids, 2012, 1-9. |
| [105] |
Karig, D. K., Iyer, S., Simpson, M. L., & Doktycz, M. J.. (2012). Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Research, 40(8), 3763-3774. |
| [106] |
Niwa, T., Kanamori, T., Ueda, T., & Taguchi, H..(2012). Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc Natl Acad Sci USA, 109, 8937-8942. |
| [107] |
Kaiser, C., Goldman, D., Tinoco, I., & Bustamante, C.. (2012). The ribosome modulates nascent protein folding. Biophysical Journal, 102(3), 68a. |
| [108] |
Wang, W., Hara, S., Liu, M., Aigaki, T., Shimizu, S., & Ito, Y.. (2011). Polypeptide aptamer selection using a stabilized ribosome display. Journal of Bioscience & Bioengineering, 112(5), 515-517. |
| [109] |
Gonza?Lez, D., Lokhande, N., Vadde, S., Zhao, Q., Cassill, A., & Renthal, R.. (2011). Luminescence resonance energy transfer in the cytoplasm of live escherichia coli cells. Biochemistry, 50(32), 6789-6796. |
| [110] |
Mallam, A. L., & Jackson, S. E.. (2011). Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nature Chemical Biology, 8(2), 147-153. |
| [111] |
Hensley, M. P., Tierney, D. L., & Crowder, M. W.. (2011). Zn(ii) binding to escherichia coli 70s ribosomes. Biochemistry, 50(46), 9937-9939. |
| [112] |
Pereira de Souza, T., Steiniger, F., Stano, P., Fahr, A., & Luisi, P. L. (2011). Spontaneous Crowding of Ribosomes and Proteins inside Vesicles: A Possible Mechanism for the Origin of Cell Metabolism. ChemBioChem, 12(15), 2325–2330. |
| [113] |
Grimm, S., Yu, F., & Nygren, P.-Å. (2011). Ribosome Display Selection of a Murine IgG1 Fab Binding Affibody Molecule Allowing Species Selective Recovery Of Monoclonal Antibodies. Molecular Biotechnology, 48(3), 263–276. |
| [114] |
Yanagida, H., Matsuura, T., Kazuta, Y., & Yomo, T. (2011). In Vitro Selection of Proteins that Undergo Covalent Labeling with Small Molecules by Thiol-Disulfide Exchange by Using Ribosome Display. ChemBioChem, 12(6), 962–969. |
| [115] |
Welsh, J. P., Bonomo, J., & Swartz, J. R.. (2011). Localization of bip to translating ribosomes increases soluble accumulation of secreted eukaryotic proteins in an escherichia coli cell-free system. Biotechnology & Bioengineering, 108(8), 1739-1748. |
| [116] |
Kihara, F., Niimi, T., Yamashita, O., & Yaginuma, T. (2011). Heat shock factor binds to heat shock elements upstream of heat shock protein 70a and Samui genes to confer transcriptional activity in Bombyx mori diapause eggs exposed to 5°C. Insect Biochemistry and Molecular Biology, 41(11), 843–851. |
| [117] |
Iizuka, R., Yamagishi-Shirasaki, M., & Funatsu, T.. (2011). Kinetic study of de novo chromophore maturation of fluorescent proteins. Biophysical Journal, 100(3), 486a. |
| [118] |
Ohtsuka, T., Neki, S., Kanai, T., Akiyoshi, K., Nomura, S. M., & Ohtsuki, T.. (2011). Synthesis and in situ insertion of a site-specific fluorescently labeled membrane protein into cell-sized liposomes. Analytical Biochemistry, 418(1), 97-101. |
| [119] |
Lam, K. N., Van Bakel, H., Cote, A. G., Anton, V. D. V., & Hughes, T. R.. (2011). Sequence specificity is obtained from the majority of modular c2h2 zinc-finger arrays. Nucleic Acids Research, 39(11), 4680-4690. |
| [120] |
De Masi, F., Grove, C. A., Vedenko, A., Alibés, A., Gisselbrecht, S. S., Serrano, L., et al. (2011). Using a structural and logics systems approach to infer bHLH–DNA binding specificity determinants. Nucleic Acids Research, 39(11), 4553–4563. |
| [121] |
Garza-Sánchez, F., Schaub, R. E., Janssen, B. D., & Hayes, C. S. (2011). tmRNA regulates synthesis of the ArfA ribosome rescue factor. Molecular Microbiology, 80(5), 1204–1219. |
| [122] |
Shingaki, T., & Nimura, N.. (2011). Improvement of translation efficiency in an escherichia coli cell-free protein system using cysteine. Protein Expression & Purification, 77(2), 193-197. |
| [123] |
Rosner, K., Kasprzak, M. F., Horenstein, A. C. J., Thurston, H. L., Abrams, J., & Kerwin, L. Y., et al. (2011). Engineering a waste management enzyme to overcome cancer resistance to apoptosis: adding dnase1 to the anti-cancer toolbox. Cancer Gene Therapy, 18(5), 346-357. |
| [124] |
Zhou, Z. P., Shimizu, Y., Tadakuma, H., Taguchi, H., Ito, K., & Ueda, T.. (2011). Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. Journal of Biochemistry, 149(5), 609-618. |
| [125] |
Chiba, S., Kanamori, T., Ueda, T., Akiyama, Y., Pogliano, K., & Ito, K. (2011). Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proceedings of the National Academy of Sciences, 108(15), 6073–6078. |
| [126] |
Yamamoto, S., Izumiya, H., Mitobe, J., Morita, M., Arakawa, E., & Ohnishi, M., et al. (2011). Identification of a chitin-induced small rna that regulates translation of the tfox gene, encoding a positive regulator of natural competence in vibrio cholerae. Journal of Bacteriology, 193(8), 1953. |
| [127] |
Subtelny, A. O., Hartman, M. C. T., & Szostak, J. W. (2011). Optimal Codon Choice Can Improve the Efficiency and Fidelity of N-Methyl Amino Acid Incorporation into Peptides by In-Vitro Translation. Angewandte Chemie International Edition, 50(14), 3164–3167. |
| [128] |
Handa, Yoshihiro, Inaho, Noriyuki, Nameki, & Nobukazu. (2011). Yaej is a novel ribosome-associated protein in escherichia coli that can hydrolyze peptidyl–trna on stalled ribosomes. Nucleic Acids Research, 39(5), 1739-1748. |
| [129] |
Ramu, H., Nora Vázquez-Laslop, Klepacki, D., Dai, Q., & Mankin, A. S.. (2011). Nascent peptide in the ribosome exit tunnel affects functional properties of the a-site of the peptidyl transferase center. Molecular cell, 41(3), 321-330. |
| [130] |
Panayiotou, C., Solaroli, N., Xu, Y., Johansson, M., & Karlsson, A.. (2011). The characterization of human adenylate kinases 7 and 8 demonstrates differences in kinetic parameters and structural organization among the family of adenylate kinase isoenzymes. Biochemical Journal, 433(3), 527. |
| [131] |
Narayan, V., Pion, E., Landre, V., Muller, P., & Ball, K. L.. (2011). Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase chip. Journal of Biological Chemistry, 286(1), 607-619. |
| [132] |
Lamichhane, T. N., Dinuka, A. N., Duc Anne-Cécile E., Cunningham, P. R., & Chow, C. S.. (2011). Selection of peptides targeting helix 31 of bacterial 16s ribosomal rna by screening m13 phage-display libraries. Molecules, 16(2), 1211-1239. |
| [133] |
Midon, M., Schafer, P., Pingoud, A., Ghosh, M., Moon, A. F., & Cuneo, M. J., et al. (2011). Mutational and biochemical analysis of the dna-entry nuclease enda from streptococcus pneumoniae. Nucleic Acids Research, 39(2), 623-634. |
| [134] |
Ma, Z., & Hartman, M. C.. (2012). In vitro selection of unnatural cyclic peptide libraries via mrna display. Methods in Molecular Biology, 805, 367-390. |
| [135] |
Yamaguchi, T., Yoshinaga, N., Yazawa, T., Gen, K., & Kitano, T.. (2010). Cortisol is involved in temperature-dependent sex determination in the japanese flounder. Endocrinology, 151(8), 3900-3908. |
| [136] |
Ueda, T.. (2010). Ribosome display with the pure technology. Methods in Molecular Biology, 607, 219-225. |
| [137] |
Kuruma, Y., Suzuki, T., & Ueda, T.. (2010). Production of multi-subunit complexes on liposome through an e. coli cell-free expression system. Methods Mol Biol, 607, 161-171. |
| [138] |
Shimizu, Y., & Ueda, T.. (2010). Pure technology. Methods in Molecular Biology, 607, 11-21. |
| [139] |
Moritani, Y., Nomura, S. I. M., Morita, I., & Akiyoshi, K.. (2010). Direct integration of cell-free-synthesized connexin-43 into liposomes and hemichannel formation. Febs Journal, 277(16), 3343-3352. |
| [140] |
Lakshmipathy, S. K., Gupta, R., Pinkert, S., Etchells, S. A., & Hartl, F. U.. (2010). Versatility of trigger factor interactions with ribosome-nascent chain complexes. Journal of Biological Chemistry, 285(36), 27911-27923. |
| [141] |
Haruichi, A., & Shaorong, C.. (2010). In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of rna polymerase holoenzyme. Nucleic Acids Research, 38(13), e141. |
| [142] |
Theerthagiri, G., Eisenhardt, N., Schwarz, H., & Antonin, W.. (2010). The nucleoporin nup188 controls passage of membrane proteins across the nuclear pore complex. The Journal of Cell Biology, 189(7), 1129-1142. |
| [143] |
Shen, B. W., Heiter, D. F., Chan, S. H., Wang, H., Xu, S. Y., & Morgan, R. D., et al. (2010). Unusual target site disruption by the rare-cutting hnh restriction endonuclease paci. Structure, 18(6), 734-743. |
| [144] |
Holmqvist, E., Reimeg?Rd, J., Sterk, M., Grantcharova, N., R?Mling, U., & Wagner, E. G. H.. (2010). Two antisense rnas target the transcriptional regulator csgd to inhibit curli synthesis. EMBO JOURNAL, 29(11), 1840-1850. |
| [145] |
Sunami, T., Hosoda, K., Suzuki, H., Matsuura, T., & Yomo, T.. (2010). Cellular compartment model for exploring the effect of the lipidic membrane on the kinetics of encapsulated biochemical reactions. Langmuir, 26(11), 8544-8551. |
| [146] |
Bonomo, J., Welsh, J. P., Manthiram, K., & Swartz, J. R.. (2010). Comparing the functional properties of the hsp70 chaperones, dnak and bip. Biophysical Chemistry, 149(1), 58-66. |
| [147] |
Nishiyama, K. I., Maeda, M., Abe, M., Kanamori, T., Shimamoto, K., & Kusumoto, S., et al. (2010). A novel complete reconstitution system for membrane integration of the simplest membrane protein. Biochemical & Biophysical Research Communications, 394(3), 733-736. |
| [148] |
Noto, T., Kurth, H. M., Kataoka, K., Aronica, L., Desouza, L. V., & Siu, K. W. M., et al. (2010). The tetrahymena argonaute-binding protein giw1p directs a mature argonaute-sirna complex to the nucleus. Cell, 140(5), 692-703. |
| [149] |
Matsumura, N., Tsuji, T., Sumida, T., Kokubo, M., Onimaru, M., & Doi, N., et al. (2010). Mrna display selection of a high-affinity, bcl-xl-specific binding peptide. The FASEB Journal, 24(7), 2201-2210. |
| [150] |
Osada, E., Shimizu, Y., Akbar, B. K., Kanamori, T., & Ueda, T.. (2009). Epitope mapping using ribosome display in a reconstituted cell-free protein synthesis system. Journal of Biochemistry, 145(5), 693-700. |
| [151] |
Tanner, D. R., Cariello, D. A., Woolstenhulme, C. J., Broadbent, M. A., & Buskirk, A. R.. (2009). Genetic identification of nascent peptides that induce ribosome stalling. Journal of Biological Chemistry, 284(50), 34809-34818. |
| [152] |
Sumida, T., Doi, N., & Yanagawa, H.. (2009). Bicistronic dna display for in vitro selection of fab fragments. Nucleic Acids Research, 37(22), e147. |
| [153] |
Eriko, M. S., Akihiko, T., Hiroyuki, T., Takuya, M., Tsutomu, N., & Tomoji, K.. (2009). Profiling of gene-dependent translational progress in cell-free protein synthesis by real-space imaging. Analytical Biochemistry, 394(2), 275-280. |
| [154] |
Yamamoto, H., Fukui, K., Takahashi, H., Kitamura, S., Shiota, T., & Terao, K., et al. (2009). Roles of tom70 in import of presequence-containing mitochondrial proteins. Journal of Biological Chemistry, 284(46), 31635-31646. |
| [155] |
Göckler, N., Jofre, G., Papadopoulos, C., Soppa, U., Tejedor, F. J., & Becker, W.. (2009). Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS Journal, 276(21), 6324–6337. |
| [156] |
Uchida, I., Ishihara, R., Tanaka, K., Hata, E., Makino, S., & Kanno, T., et al. (2009). Salmonella enterica serotype typhimurium dt104 arta-dependent modification of pertussis toxin-sensitive g proteins in the presence of [32p]nad. Microbiology, 155(11), 3710-3718. |
| [157] |
Feng, Y., & Cronan, J. E.. (2009). A new member of the escherichia coli fad regulon: transcriptional regulation of fadm (ybaw). Journal of Bacteriology, 191(20), 6320-6328. |
| [158] |
Solaroli, N., Panayiotou, C., Johansson, M., & Karlsson, A.. (2009). Identification of two active functional domains of human adenylate kinase 5. Febs Letters, 583(17), 2872-2876. |
| [159] |
Pfeiffer, V., Papenfort, K., Lucchini, S., Hinton, J. C. D., & Vogel, J.. (2009). Coding sequence targeting by micc rna reveals bacterial mrna silencing downstream of translational initiation. NATURE STRUCTURAL & MOLECULAR BIOLOGY, 16(8), 840-846. |
| [160] |
Estevez-Torres, A., Crozatier, C., Diguet, A., Hara, T., Saito, H., & Yoshikawa, K., et al. (2009). Sequence-independent and reversible photocontrol of transcription/expression systems using a photosensitive nucleic acid binder. Proceedings of the National Academy of Sciences, 106(30), 12219-12223. |
| [161] |
Estevez-Torres, A., Crozatier, C., Diguet, A., Hara, T., Saito, H., & Yoshikawa, K., et al. (2009). Sequence-independent and reversible photocontrol of transcription/expression systems using a photosensitive nucleic acid binder. Proceedings of the National Academy of Sciences, 106(30), 12219-12223. |
| [162] |
Takahashi, S., Iida, M., Furusawa, H., Shimizu, Y., Ueda, T., & Okahata, Y.. (2009). Real-time monitoring of cell-free translation on a quartz-crystal microbalance. Journal of the American Chemical Society, 131(26), 9326-9332. |
| [163] |
Kuroha, K., Horiguchi, N., Aiba, H., & Inada, T. (2009). Analysis of nonstop mRNA translation in the absence of tmRNA inEscherichia coli. Genes to Cells, 14(6), 739–749. |
| [164] |
Osada, E., Shimizu, Y., Akbar, B. K., Kanamori, T., & Ueda, T.. (2009). Epitope mapping using ribosome display in a reconstituted cell-free protein synthesis system. Journal of Biochemistry, 145(5), 693-700. |
| [165] |
Niwa, T., Ying, B. W., Saito, K., Jin, W., Takada, S., & Ueda, T., et al. (2009). Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of escherichia coli proteins. Proceedings of the National Academy of Sciences, 106(11), 4201-4206. |
| [166] |
Robin, Togashi, S., Ryder, D. M., Wall, A. G., & J., G.. (2009). Trigger factor from the psychrophilic bacterium psychrobacter frigidicola is a monomeric chaperone. Journal of Bacteriology, 191(4), 1162-1168. |
| [167] |
Matsuura, T., Kazuta, Y., Aita, T., Adachi, J., & Yomo, T.. (2009). Quantifying epistatic interactions among the components constituting the protein translation system. Molecular Systems Biology, 5(1). |
| [168] |
Zheng, Y., Posfai, J., Morgan, R. D., Vincze, T., & Roberts, R. J.. (2009). Using shotgun sequence data to find active restriction enzyme genes. Nucleic Acids Research, 37(1), e1. |
| [169] |
Hosoda, K., Sunami, T., Kazuta, Y., Matsuura, T., Suzuki, H., & Yomo, T.. (2008). Quantitative study of the structure of multilamellar giant liposomes as a container of protein synthesis reaction. Langmuir, 24(23), 13540-13548. |
| [170] |
Terashima, H., Abe-Yoshizumi, R., Kojima, S., & Homma, M.. (2008). Cell-free synthesis of the torque-generating membrane proteins, poma and pomb, of the na+-driven flagellar motor in vibrio alginolyticus. Journal of Biochemistry, 144(5), 635-642. |
| [171] |
Kazuta, Y., Adachi, J., Matsuura, T., Ono, N., Mori, H., & Yomo, T.. (2008). Comprehensive analysis of the effects of escherichia coli orfs on protein translation reaction. Molecular & Cellular Proteomics, 7(8), 1530-1540. |
| [172] |
Maki, K., Uno, K., Morita, T., & Aiba, H.. (2008). Rna, but not protein partners, is directly responsible for translational silencing by a bacterial hfq-binding small rna. Proceedings of the National Academy of Sciences, 105(30), 10332-10337. |
| [173] |
Uemura, S., Iizuka, R., Ueno, T., Shimizu, Y., Taguchi, H., & Ueda, T., et al. (2008). Single-molecule imaging of full protein synthesis by immobilized ribosomes. Nucleic Acids Research, 36(12), e70. |
| [174] |
Uemura, S., Iizuka, R., Ueno, T., Shimizu, Y., Taguchi, H., & Ueda, T., et al. (2008). Single-molecule imaging of full protein synthesis by immobilized ribosomes. Nucleic Acids Research, 36(12), e70. |
| [175] |
Sako, Y., Morimoto, J., Murakami, H., & Suga, H.. (2008). Ribosomal synthesis of bicyclic peptides via two orthogonal inter-side-chain reactions. Journal of the American Chemical Society, 130(23), 7232-7234. |
| [176] |
Vazquezlaslop, N., Thum, C., & Mankin, A. S.. (2008). Molecular mechanism of drug-dependent ribosome stalling. Molecular Cell, 30(2), 190-202. |
| [177] |
Sako, Y., Goto, Y., Murakami, H., & Suga, H.. (2008). Ribosomal synthesis of peptidase-resistant peptides closed by a nonreducible inter-side-chain bond. ACS Chemical Biology, 3(4), 241-249. |
| [178] |
Urban, J. H., & Vogel, J.. (2008). Two seemingly homologous noncoding rnas act hierarchically to activate glms mrna translation. PLoS Biology, 6(3), e64. |
| [179] |
Ozaki, Y., Suzuki, T., Kuruma, Y., Ueda, T., & Yoshida, M.. (2008). Unci protein can mediate ring-assembly of c-subunits of fof1-atp synthase in vitro. Biochemical & Biophysical Research Communications, 367(3), 663-666. |
| [180] |
Sakamoto, A., Yamagishi, M., Watanabe, T., Aizawa, Y., Kato, T., & Funatsu, T.. (2008). Fluorescence labeling of a cytokine with desthiobiotin-tagged fluorescent puromycin. Journal of Bioscience & Bioengineering, 105(3), 238-242. |
| [181] |
Goto, Y., Ohta, A., Sako, Y., Yamagishi, Y., Murakami, H., & Suga, H.. (2008). Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS Chemical Biology, 3(2), 120-129. |
| [182] |
Kawakami, T., Murakami, H., & Suga, H.. (2008). Messenger rna-programmed incorporation of multiple n-methyl-amino acids into linear and cyclic peptides. Chemistry & Biology, 15(1), 32-42. |
| [183] |
Neely, R. K., & Roberts, R. J.. (2008). The BsaHI restriction-modification system: Cloning, sequencing and analysis of conserved motifs. BMC Molecular Biology, 9(1), 48. |
| [184] |
Hillebrecht, J. R., & Chong, S.. (2008). A comparative study of protein synthesis in in vitro systems: from the prokaryotic reconstituted to the eukaryotic extract-based. BMC Biotechnology, 8(1), 58. |
| [185] |
Yanagida, H., Matsuura, T., & Yomo, T.. (2008). Compensatory evolution of a ww domain variant lacking the strictly conserved trp residue. Journal of Molecular Evolution, 66(1), 61-71. |
| [186] |
Ohta, A., Murakami, H., Higashimura, E., & Suga, H.. (2007). Synthesis of polyester by means of genetic code reprogramming. Chemistry & Biology (Cambridge), 14(12), 1315-1322. |
| [187] |
Doi, Y., Ohtsuki, T., Shimizu, Y., Ueda, T., & Sisido, M.. (2007). Elongation factor tu mutants expand amino acid tolerance of protein biosynthesis system. Journal of the American Chemical Society, 129(46), 14458-14462. |
| [188] |
Murtas, G., Kuruma, Y., Bianchini, P., Diaspro, A., & Luisi, P. L. (2007). Protein synthesis in liposomes with a minimal set of enzymes. Biochemical and Biophysical Research Communications, 363(1), 12–17. |
| [189] |
Sharma, C. M., Darfeuille, F., Plantinga, T. H., & Vogel, J.. (2007). A small rna regulates multiple abc transporter mrnas by targeting c/a-rich elements inside and upstream of ribosome-binding sites. Genes & Development, 21(21), 2804-2817. |
| [190] |
Kojima, K., Oshita, M., Nanjo, Y., Kasai, K., Tozawa, Y., Hayashi, H., & Nishiyama, Y. (2007). Oxidation of elongation factor G inhibits the synthesis of the D1 protein of photosystem II. Molecular Microbiology, 65(4), 936–947. |
| [191] |
Sando, S., Abe, K., Sato, N., Shibata, T., Mizusawa, K., & Aoyama, Y. (2007). Unexpected Preference of theE. coliTranslation System for the Ester Bond during Incorporation of Backbone-Elongated Substrates. Journal of the American Chemical Society, 129(19), 6180–6186. |
| [192] |
Lakshmipathy, S. K., Tomic, S., Kaiser, C. M., Chang, H. C., Genevaux, P., & Georgopoulos, C., et al. (2007). Identification of nascent chain interaction sites on trigger factor. Journal of Biological Chemistry, 282(16), 12186-12193. |
| [193] |
Matsuura, T., Yanagida, H., Ushioda, J., Urabe, I., & Yomo, T. (2007). Nascent chain, mRNA, and ribosome complexes generated by a pure translation system. Biochemical and Biophysical Research Communications, 352(2), 372–377. |
| [194] |
Ohashi, H., Shimizu, Y., Ying, B. W., & Ueda, T.. (2007). Efficient protein selection based on ribosome display system with purified components. Biochemical & Biophysical Research Communications, 352(1), 270-276. |
| [195] |
Udagawa, T., Shimizu, Y., & Ueda, T.. (2004). Evidence for the translation initiation of leaderless mrnas by the intact 70 s ribosome without its dissociation into subunits in eubacteria. Journal of Biological Chemistry, 279(10), 8539-8546. |
| [196] |
Ueno, S., Arai, H., Suzuki, M., & Husimi, Y.. (2007). An mrna-protein fusion at n-terminus for evolutionary protein engineering. International Journal of Biological Sciences, 3(6), 365-374. |
| [197] |
Narita, A., Ogawa, K., Sando, S., & Aoyama, Y.. (2007). cis-regulatory hairpin-shaped mrna encoding a reporter protein: catalytic sensing of nucleic acid sequence at single nucleotide resolution. NATURE PROTOCOLS, 2(5), 1105-1116. |
| [198] |
Yoshimori, A., Sakai, J., Sunaga, S., Kobayashi, T., & Tanuma, S. I.. (2007). Structural and functional definition of the specificity of a novel caspase-3 inhibitor, ac-dnld-cho. BMC Pharmacology, 7(1), 8. |
| [199] |
Kawahashi, Y., Doi, N., Oishi, Y., Tsuda, C., Takashima, H., & Baba, T., et al. (2006). High-throughput fluorescence labelling of full-length cdna products based on a reconstituted translation system. Journal of Biochemistry, 141(1), 19-24. |
| [200] |
Saguy, M., Gillet, R., Skorski, P., Hermann-Le Denmat, S., & Felden, B.. (2007). Ribosomal protein s1 influences trans-translation in vitro and in vivo. Nucleic Acids Research, 35(7), 2368-2376. |
| [201] |
Zheng, Y., & Roberts, R. J.. (2007). Selection of restriction endonucleases using artificial cells. Nucleic Acids Research, 35(11), e83. |
| [202] |
Setoguchi, K., Otera, H., & Mihara, K.. (2006). Cytosolic factor- and tom-independent import of c-tail-anchored mitochondrial outer membrane proteins. EMBO JOURNAL, 25(24), 5635-5647. |
| [203] |
Sunami, T., Sato, K., Matsuura, T., Tsukada, K., Urabe, I., & Yomo, T.. (2006). Femtoliter compartment in liposomes for in vitro selection of proteins. Analytical Biochemistry, 357(1), 128-136. |
| [204] |
Ying, & B.-W. (2006). Co-translational binding of groel to nascent polypeptides is followed by post-translational encapsulation by groes to mediate protein folding. Journal of Biological Chemistry, 281(31), 21813-21819. |
| [205] |
Ishihara, N., Fujita, Y., Oka, T., & Mihara, K.. (2006). Regulation of mitochondrial morphology through proteolytic cleavage of opa1. EMBO JOURNAL, 25(13), 2966-2977. |
| [206] |
Groves, M., Lane, S.,Douthwaite, J., Lowne, D., Rees, D. G., & Edwards, B., et al. (2012). Affinity maturation of phage display antibody populations using ribosome display. Methods in Molecular Biology, 313(1), 129-139. |
| [207] |
Villemagne, D., Jackson, R., & Douthwaite, J. A.. (2006). Highly efficient ribosome display selection by use of purified components for in vitro translation. Journal of Immunological Methods, 313(1-2), 140-148. |
| [208] |
Yamamoto, T., Izumi, S., & Gekko, K.. (2006). Mass spectrometry of hydrogen/deuterium exchange in 70s ribosomal proteins from e. coli. Febs Letters, 580(15), 0-3642. |
| [209] |
Shimizu, Y., & Ueda, T.. (2006). Smpb triggers gtp hydrolysis of elongation factor tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. Journal of Biological Chemistry, 281(23), 15987-15996. |
| [210] |
Seebeck, F. P., & Szostak, J. W.. (2006). Ribosomal synthesis of dehydroalanine-containing peptides. Journal of the American Chemical Society, 128(22), 7150-7151. |
| [211] |
Kubota, S., Kubota, H., & Nagata, K.. (2006). Cytosolic chaperonin protects folding intermediates of gβ from aggregation by recognizing hydrophobic β-strands. Proceedings of the National Academy of Sciences of the United States of America, 103(22), 8360-8365. |
| [212] |
Muto, H., Nakatogawa, H., & Ito, K.. (2006). Genetically encoded but nonpolypeptide prolyl-trna functions in the a site for secm-mediated ribosomal stall. Molecular Cell, 22(4), 545-552. |
| [213] |
Murakami, H., Ohta, A., Ashigai, H., & Suga, H.. (2006). A highly flexible trna acylation method for non-natural polypeptide synthesis. Nature Methods, 3(5), 357-359. |
| [214] |
Umekage, S., & Ueda, T.. (2006). Spermidine inhibits transient and stable ribosome subunit dissociation. Febs Letters, 580(5), 0-1226. |
| [215] |
Itoh, H., Kawazoe, Y., & Shiba, T.. (2006). Enhancement of protein synthesis by an inorganic polyphosphate in an e. coli cell-free system. Journal of Microbiological Methods, 64(2), 241-249. |
| [216] |
Ogawa, A., Sando, S., & Aoyama, Y.. (2010). Termination‐free prokaryotic protein translation by using anticodon‐adjusted e. coli trnaser as unified suppressors of the uaa/uga/uag stop codons. read‐through ribosome display of full‐length dhfr with translated utr as a buried spacer arm. Chembiochem, 7(2), 249-252. |
| [217] |
Tomic, S., Johnson, A. E., Hartl, F. U., & Etchells, S. A. (2005). Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Letters, 580(1), 72–76. |
| [218] |
Hallier, M. (2006). Small protein B interacts with the large and the small subunits of a stalled ribosome during trans-translation. Nucleic Acids Research, 34(6), 1935–1943. |
| [219] |
Jarutat, T., Frisch, C., Nickels, C., Merz, H., & Knappik, A.. (2006). Isolation and comparative characterization of ki-67 equivalent antibodies from the hucal? phage display library. Biological Chemistry, 387(7). |
| [220] |
Josephson, K., Hartman, M. C. T., & Szostak, J. W. (2005). Ribosomal Synthesis of Unnatural Peptides. Journal of the American Chemical Society, 127(33), 11727–11735. |
| [221] |
Shimizu, Y., Kanamori, T., & Ueda, T.. (2005). Protein synthesis by pure translation systems. Methods (Amsterdam), 36(3), 299-304. |
| [222] |
Sando, S., Kanatani, K., Sato, N., Matsumoto, H., Hohsaka, T., & Aoyama, Y.. (2005). A small-molecule-based approach to sense codon-templated natural-unnatural hybrid peptides. selective silencing and reassignment of the sense codon by orthogonal reacylation stalling at the single-codon level. Journal of the American Chemical Society, 127(22), 7998-7999. |
| [223] |
Fukushima, K., Ikehara, Y., & Yamashita, K. (2005). Functional Role Played by the Glycosylphosphatidylinositol Anchor Glycan of CD48 in Interleukin-18-induced Interferon-γ Production. Journal of Biological Chemistry, 280(18), 18056–18062. |
| [224] |
Yano, M., Okano, H. J., & Okano, H.. (2005). Involvement of hu and heterogeneous nuclear ribonucleoprotein k in neuronal differentiation through p21 mrna post-transcriptional regulation. Journal of Biological Chemistry, 280(13), 12690-12699. |
| [225] |
Ying, B. W., Taguchi, H., Kondo, M., & Ueda, T.. (2005). Co-translational involvement of the chaperonin groel in the folding of newly translated polypeptides. Journal of Biological Chemistry, 280(12), 12035-12040. |
| [226] |
Tokunaga, M., Mizukami, M., & Tanaka, R.. (2005). Novel processing and localization of cata, ccda associated thiol-disulfide oxidoreductase, in protein hyper-producing bacterium brevibacillus choshinensis. Protein & Peptide Letters, 12(1), 95-98. |
| [227] |
Kuruma, Y., Nishiyama, K. I., Shimizu, Y., Matthias Müller, & Ueda, T.. (2005). Development of a minimal cell-free translation system for the synthesis of presecretory and integral membrane proteins. Biotechnology progress, 21(4), 1243-1251. |
| [228] |
Ying, B.-W., Taguchi, H., Ueda, H., & Ueda, T. (2004). Chaperone-assisted folding of a single-chain antibody in a reconstituted translation system. Biochemical and Biophysical Research Communications, 320(4), 1359–1364. |
| [229] |
Asai, T., Takahashi, T., Esaki, M., Nishikawa, S. I., Ohtsuka, K., & Nakai, M., et al. (2004). Reinvestigation of the requirement of cytosolic atp for mitochondrial protein import. Journal of Biological Chemistry, 279(19), 19464-19470. |
| [230] |
Kawano, M., Suzuki, S., Suzuki, M., Oki, J., & Imamura, T.. (2004). Bulge- and basal layer-specific expression of fibroblast growth factor-13 (fhf-2) in mouse skin. Journal of Investigative Dermatology, 122(5), 1084-1090. |