上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
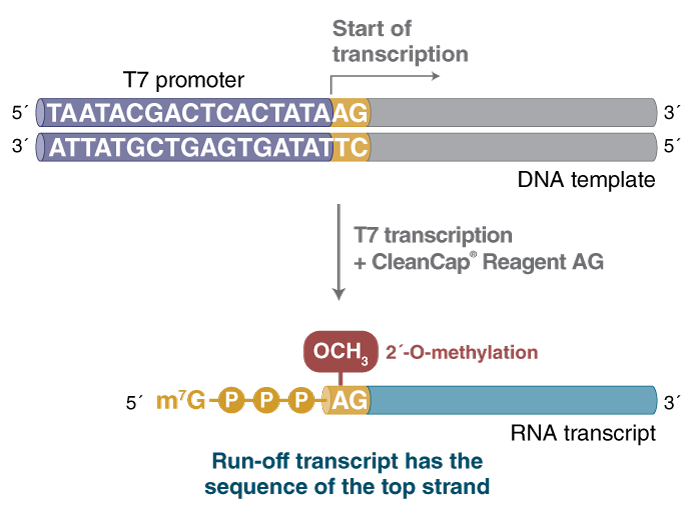
Most eukaryotic mRNAs require a 7-methyl guanosine (m7G) cap structure at the 5´-end and a poly(A) tail at the 3′-end for efficient translation. The HiScribe T7 mRNA Kit with CleanCap Reagent AG utilizes an optimized RNA synthesis formulation and trinucleotide cap analog technology for co-transcriptionally capping mRNAs that contain a natural Cap-1 structure in a single simplified reaction without compromising RNA yield. By using a DNA template with a T7 promoter sequence followed by an AG initiation sequence and an encoded poly(A) tail, mRNAs can be transcribed with a 5´-m7G Cap-1 structure that is polyadenylated, translationally competent and able to evade the cellular innate immune response.
The HiScribe T7 mRNA Kit with CleanCap Reagent AG is formatted with individual vials of NTPs and CleanCap Reagent AG to allow for partial or complete substitution of modified NTPs, with a total kit yield of 1.8 mg of mRNA. Cap-1 mRNA synthesized from this kit is suitable for many applications, including transfections, microinjections, in vitro translation, preclinical mRNA therapeutic mRNA studies as well as RNA structure and function analysis.
This kit contains sufficient reagents for 20 reactions (20 µl each). Each standard reaction yields ≥ 90 µg of RNA from 1 µg CLuc AG Control Template DNA. Each kit can yield ≥ 1.8 mg RNA.


- 产品类别:
- RNA Capping,
- RNA Synthesis In vitro Transcription (IVT)
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E2080S -20 T7 RNA Polymerase Mix M0255AAVIAL -20 1 x 0.04 ml Not Applicable LiCl Solution B2051AVIAL -20 1 x 1.4 ml Not Applicable DNase I (RNase-free) M0303AAVIAL -20 1 x 0.04 ml 2,000 units/ml CLuc AG Control Template N2078AVIAL -20 1 x 0.02 ml 0.25 mg/ml 10X T7 CleanCap® Reagent AG Reaction Buffer B2082AVIAL -20 1 x 0.04 ml Not Applicable CleanCap® Reagent AG S1413AVIAL -20 1 x 0.04 ml 40 mM ATP N0489AVIAL -20 1 x 0.04 ml 60 mM GTP N0490AVIAL -20 1 x 0.04 ml 50 mM CTP N0491AVIAL -20 1 x 0.04 ml 50 mM UTP N0492AVIAL -20 1 x 0.04 ml 50 mM Dithiothreitol (DTT) B1222AVIAL -20 1 x 0.5 ml 100 mM
-
-
特性和用法
需要但不提供的材料
DNA Template:
The DNA template must be linear and contain the T7 RNA Polymerase Promoter with the correct orientation in relation to the target sequence to be transcribed, followed by an AG initiation sequence.Modified NTPs:
Biotin-, Fluorescein-, Digoxigenin-, Aminoallyl-, Pseudouridine-5—Triphosphate, etc.General:
Thermocycler, microcentrifuge, nuclease-free water, nuclease-free tubes and tipsPurification:
Phenol, chloroform, ethanol, 3 M Sodium Acetate, pH 5.2 or Ammonium Acetate, Monarch® RNA Cleanup Kit (500 µg; T2050), equipment and reagents for RNA quantitationGel Analysis:
Gels, running buffers, loading dye, nucleic acid ladders, gel apparatus, power supply
-
优势和特性
Features
- Streamlined workflow with single-step co-transcriptional capping
- CleanCap® Reagent AG trinucleotide cap technology results in a natural Cap-1 structure, maximizing translatability and minimizing immune response from synthetic mRNA
- High capping efficiency (> 95% capped material1)
- Optimized for high yields
- Suitable for full- or partial- modified nucleotide substitution
1Final capping is dependent upon the CleanCap Reagent, DNA template and final mRNA sequence. Secondary structure due to RNA length and base composition can affect final capping efficiency.
-
相关产品
相关产品
- e2040-hiscribe-t7-high-yield-rna-synthesis-kit
- e2050-hiscribe-t7-quick-high-yield-rna-synthesis-kit
- e2060-hiscribe-t7-arca-mrna-kit-with-tailing
- 2X RNA 上样染料
- m0307-rnase-inhibitor-human-placenta
- 小鼠 RNase 抑制剂
- m0303-dnase-i-rnase-free
- ssRNA Ladder
- 低分子量 ssRNA Ladder
- E. coli Poly(A) 聚合酶
- T2050 Monarch RNA Cleanup Kit 500 ug
- T2040 Monarch RNA Cleanup Kit 50 ug
- Q5® 热启动超保真 DNA 聚合酶
- Q5® 热启动超保真 2X 预混液
- Monarch® PCR & DNA 纯化试剂盒(5 μg)
- Q5® 定点突变试剂盒
- Q5® 定点突变试剂盒(不含感受态细胞)
- NEB® 5-alpha E. coli 感受态细胞(高效级)
- SOC Outgrowth 培养基
- t1010-monarch-plasmid-miniprep-kit
- e2065-hiscribe-t7-arca-mrna-kit
操作说明、说明书 & 用法
-
操作说明
- Standard RNA Synthesis Protocol using the HiScribe T7 mRNA Kit with CleanCap Reagent AG (NEB #E2080)
- mRNA Synthesis Protocol with Modified Nucleotides using the HiScribe T7 mRNA Kit with CleanCap Reagent AG (NEB #E2080)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE2080
-
应用实例
- Scaling of High-Yield In vitro Transcription Reactions for Linear Increase of RNA Production
工具 & 资源
-
选择指南
- Recommended HiScribe® RNA Synthesis Kits by Application
FAQs & 问题解决指南
-
FAQs
- Do I need to change my promoter sequence to use CleanCap® Reagent AG?
- How can I change the initiating sequence of my dsDNA template?
- Will the 5′ base of my RNA be an “A”?
- Can an uncut plasmid be used as a template for the HiScribe® T7 mRNA Kit with CleanCap® Reagent AG?
- Can modified nucleotides be used with the HiScribe® T7 mRNA Kit with CleanCap® Reagent AG?
- How can I improve the yield of RNA when using the HiScribe® T7 mRNA Kit with CleanCap® Reagent AG?
- How can I purify the RNA synthesized using this kit?
- Can components from other HiScribe® kits, T7 RNA Polymerase (NEB #M0251) or RNAPol Reaction Buffer (NEB #B9012) be substituted in this kit?
- Do you recommend template-encoding the poly(A) tail or using E. coli Poly(A) Polymerase?
- Can I use a different CleanCap® Reagent analog with this kit?
- Are modified nucleotides included in the kit?
- Do I need to add DTT to the reaction?
-
问题解决指南
Verify that the sequence following the T7 promoter contains the AG initiating sequence.
Control Reaction
The CLuc AG control template is a linearized plasmid containing the Cypridina luciferase gene under the transcriptional control of the T7 promoter. The initiating sequence has been changed to an AG by site-directed mutagenesis to be compatible with CleanCap Reagent AG. The size of the run-off transcript is ~1.76 kb. The control reaction, following the standard reaction protocol, should yield > 90 µg of RNA in 2 hours at 37°C. If the control reaction is not working, there may be technical issues with the reaction set up. Repeat the reaction following the protocol exactly (thawing specified reagents to room temperature, setting the reaction up at room temperature, and adding the components in the exact order listed in the manual. Take every precaution to avoid RNase contamination. The control plasmid sequence can be found within the DNA Sequences and Maps Tool under the name “pCMV-CLuc2 AG Control Plasmid”. The CLuc AG control template is generated by linearizing the plasmid with the restriction enzyme XbaI.
Low Yield of Full-length RNA
If the transcription reaction generates full-length RNA but yields are significantly lower than expected, it is possible that contaminants in the DNA template are inhibiting the RNA Polymerase or the DNA template concentration may be incorrect or too low. Additional purification of the DNA template may be required. Phenol:chloroform extraction is recommended (see template DNA preparation section).
Low yield of Short Transcript
High yields of short transcripts (< 300 nts) are achieved by extending incubation time and increasing the amount of template. Incubation of reactions up to 16 hours (overnight) or using up to 2 μg of template DNA can help achieve maximum yield.
RNA Transcript Smearing on Denaturing Gel
If the RNA appears degraded (smeared) on a denaturing agarose or polyacrylamide gel, the DNA template may be contaminated with RNase. DNA templates contaminated with RNase can affect the length and yield of RNA synthesized (smear below the expected RNA length). If the DNA template is contaminated with RNase, we recommend performing phenol:chloroform extraction followed by ethanol precipitation (see template DNA preparation section) and dissolving the DNA in nuclease-free water.
RNA Transcript of Larger Size than Expected
If the RNA transcript appears larger than the expected size on a denaturing gel when compared to a single-stranded RNA ladder, the plasmid DNA that is used for the template may not be completely digested. Even if small amounts of undigested circular plasmid DNA is present, T7 RNA Polymerase can produce large amounts of long transcripts. Check the digestion of the plasmid for complete digestion compared to a sample of undigested plasmid. If undigested plasmid is present repeat the restriction digest. Alternatively, larger sized bands may be observed when the RNA is not completely denatured due to the presence of strong secondary structure.
RNA Transcript of Smaller Size than Expected
If denaturing gel analysis indicates the presence of smaller bands than expected it is most likely due to premature termination by T7 RNA Polymerase. Sequences that resemble T7 RNA Polymerase termination signals will cause premature termination. For GC-rich templates, or templates with known strong secondary structure, incubation at 42°C may improve the yield of full-length transcript.
