上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
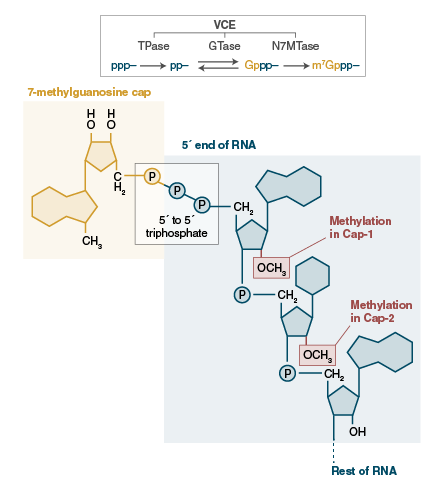
Based on the Vaccinia virus Capping Enzyme (VCE), the Vaccinia Capping System provides the necessary components to add 7-methylguanylate cap structures (Cap-0) to the 5´end of RNA (1). In eukaryotes, these terminal cap structures are involved in stabilization (2), transport (3), and translation (4) of mRNAs. Enzymatic production of capped RNA is an easy way to improve the stability and translational competence of RNA used for in vitro translation, transfection, and microinjection. Alternatively, use of labeled GTP in a reaction provides a convenient way to label any RNA containing a 5´ terminal triphosphate.
Vaccinia capping enzyme is composed of two subunits (D1 and D12). The D1 subunit carries three enzymatic activities (RNA triphosphatase, guanylyltransferase and guanine methyltransferase); all necessary for addition of a complete Cap-0 structure, m7Gppp5’N to 5′ triphosphate RNA (5,6). In vitro transcripts can be capped in less than one hour in the presence of the capping enzyme, reaction buffer, GTP, and the methyl donor, SAM. Capping is nearly 100% efficient and all capped structures are added in the proper orientation, unlike co-transcriptional addition of some cap analogs (7).
产品来源
An E. coli strain that carries an engineered His-tagged Vaccine capping gene.
- 产品类别:
- RNA Capping
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
M2080S -20 Vaccinia Capping System M2080SVIAL -20 1 x 0.04 ml 10,000 units/ml Capping Buffer B2080AVIAL -20 1 x 0.1 ml Not Applicable S-adenosylmethionine (SAM) B9003SVIAL -20 1 x 0.1 ml 32 mM GTP N2080AVIAL -20 1 x 0.05 ml 10 mM
-
-
特性和用法
单位定义
One unit of Vaccinia Capping Enzyme is defined as the amount of enzyme required to incorporate 10 pmol of (α32P) GTP into an 80 nt transcript in 1 hour at 37°C.
反应条件
1X Capping Buffer
Supplement with 0.5 mM GTP 和 0.1 mM S-腺苷甲硫氨酸(SAM)
Incubate at 37°C1X Capping Buffer
50 mM Tris-HCl
5 mM KCl
1 mM MgCl2
1 mM DTT
(pH 8 @ 25°C)贮存溶液
20 mM Tris-HCl
100 mM NaCl
1 mM DTT
0.1 mM EDTA
50% Glycerol
0.1% (w/v) Triton® X-100
pH 8 @ 25°C -
优势和特性
应用特性
Capping mRNA prior to translation assays/in vitro translation
Labeling 5´ end of mRNA -
相关产品
单独销售的组分
- S-腺苷甲硫氨酸(SAM)
- mRNA 帽结构 2-O-甲基转移酶
-
注意事项
- (建立反应体系前请仔细阅读)
RNA 在加帽前应进行纯化,并将 RNA 重悬于无核酶水中。溶液中不应存在 EDTA 和盐。
- 虽然 RNase 抑制剂不是必需添加的,但许多用户更喜欢使用它来提高溶液中 RNA 的稳定性。如有需要,可以在建立反应体系时添加 0.5 µl 标准 RNase 抑制剂(如小鼠 RNase 抑制剂,NEB #M0314)。加帽和标记实验流程第 1 步中所用的 H2O 量相应减少体积即可。
- 在与牛痘病毒加帽酶温育前,加热 RNA 溶液,可去除转录本 5´ 末端的二级结构。对于已知 5´ 末端高度结构化的转录本,将时间延长至 10 分钟。
- SAM 在 pH 值为 7 – 8 和 37℃ 条件下不稳定,应在开始反应前再配置新鲜混合液。建议首先确定要建立的反应次数,然后在快要建立反应体系时将 32 mM 贮存液分装稀释至 2 mM。应将该“工作原液”放置于冰上,防止 SAM 降解。
- 对于已知具有 5´ 末端结构化的转录本,可将反应时间延长至 60 分钟来提高加帽效率。
- 标记 5´ 末端时,GTP 总浓度应为反应中 mRNA 摩尔浓度的 1–3 倍。可稀释 10 mM 的贮存液,并掺入标记的 GTP 制备 GTP 混合液。
- (建立反应体系前请仔细阅读)
-
参考文献
- Shuman, S. (1990). J. Biol. Chem. 265, 11960-11966.
- Furiichi, Y. et al (1977). Nature. 266, 235-239.
- Lewis, J.D. and Izaurralde, E (1997). Eur. J. Biochem. 247, 461-469.
- Iizuka, N. et al. (1994). Mol. Cell. Biol. 14, 7322-7330.
- Guo, P. and Moss, B. (1990). Proc. Natl. Acad. Sci. 87, 4023-4027.
- Mao, X. and Shuman, S. (1994). J. Biol. Chem. 269, 24472-24479.
- Grudzien, E. et al. (2004). RNA. 10, 1479.
- Ramanathan, A. et al. (2016). Nucleic Acids Res. Sep 19; 44(16), 7511–7526. PubMedID: 27317694
操作说明、说明书 & 用法
-
操作说明
- Labeling Protocol (M2080)
- Cap-0 synthesis using Vaccinia Capping Enzyme (NEB #M2080)
- Poly(A) Tailing of RNA using E. coli Poly(A) Polymerase (NEB# M0276)
- Cappable-seq for prokaryotic transcription start site (TSS) determination
- One-Step Cap-1 mRNA synthesis with Vaccinia Capping Enzyme (VCE) and mRNA Cap 2´-O-methyltransferase (2´-O-me) (NEB #M0366, #M2080)
-
应用实例
- Scaling of High-Yield In vitro Transcription Reactions for Linear Increase of RNA Production
FAQs & 问题解决指南
-
FAQs
- What is Cap-0 and Cap-1?
- Can Vaccinia Capping Enzyme (NEB #M2080) use cap analogs in capping reaction?
-
实验技巧
- It is important to ensure that the RNA is purified prior to use. It should be suspended in nuclease-free water free from EDTA and salts.
- Heating the RNA at 65°C for 5 minutes prior to setting up the reaction removes secondary structures on the 5´ end of the transcript. Extend time to 10 minutes for RNA with known highly structured 5’ ends.
- SAM is unstable at pH 7–8, 37°C. Hence, it should be diluted just prior to starting the reaction.
- We strongly recommend wearing gloves, using nuclease-free tubes and reagents, and thoroughly cleaning pipettes and bench surfaces to avoid RNase contamination.
