上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
Download the NEBNext UltraExpress FS DNA Library Prep Kit Data Supplement.
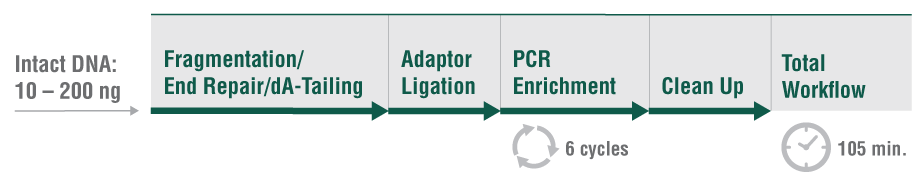
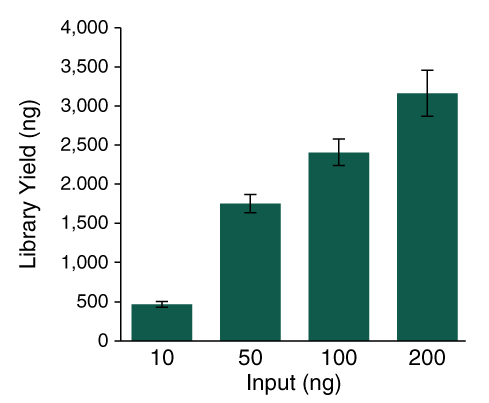
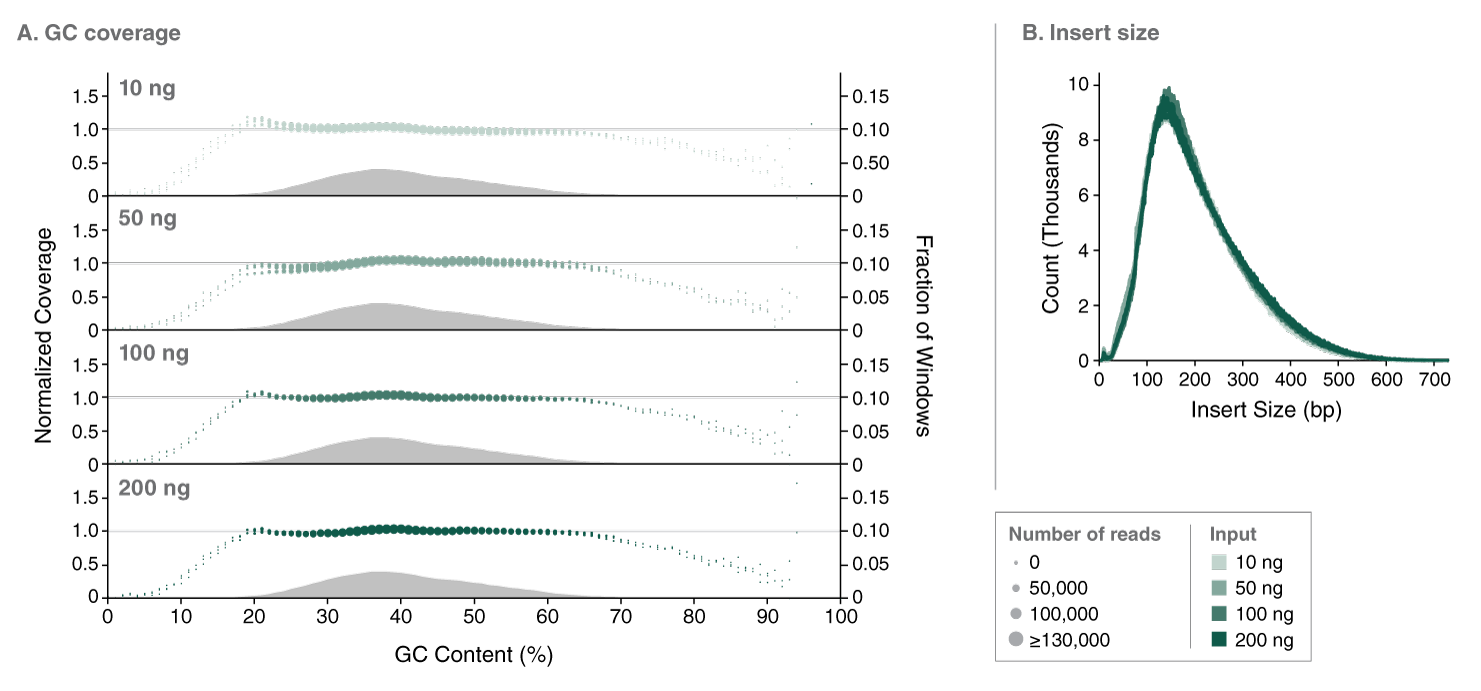
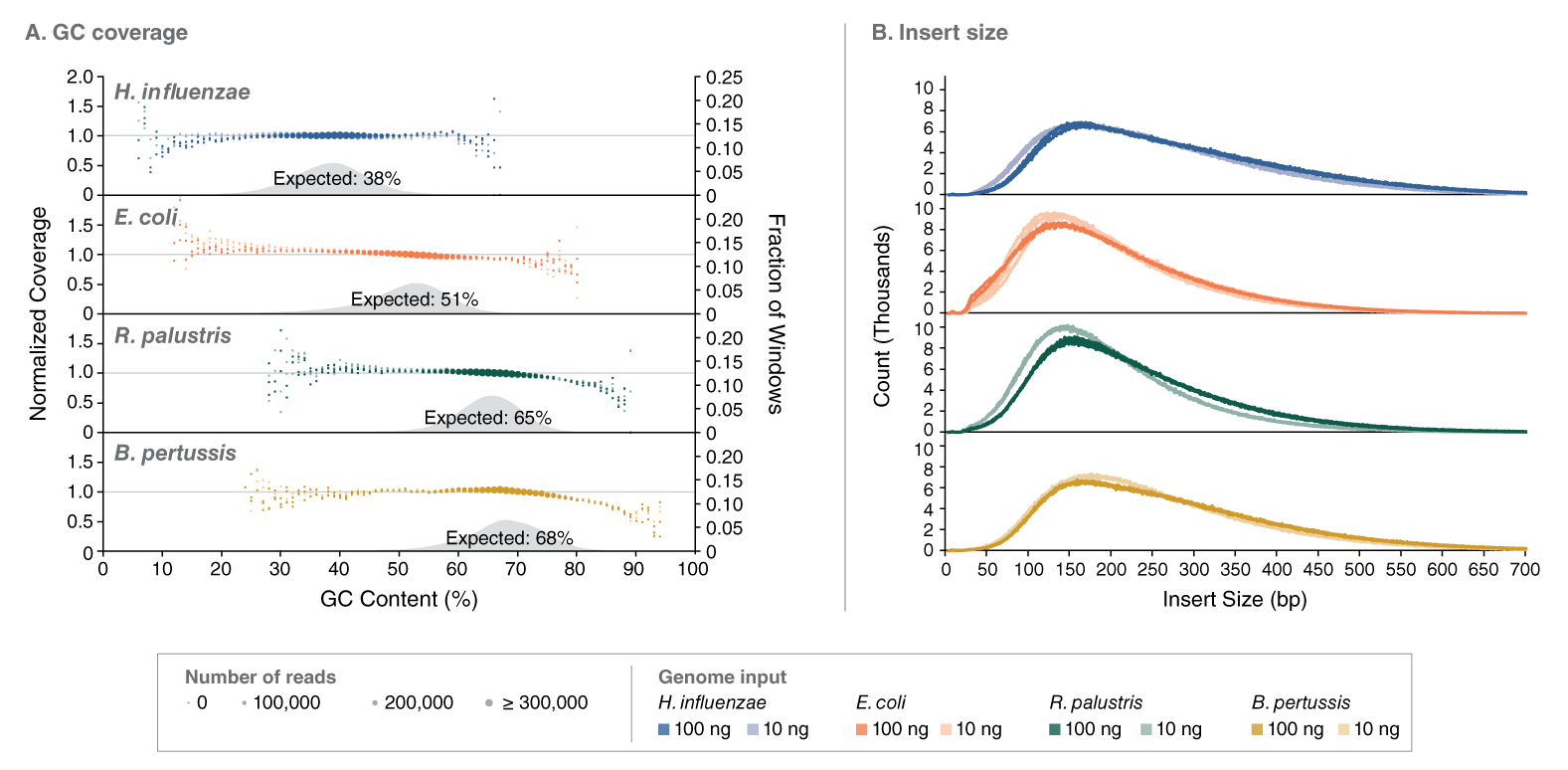
The NEBNext UltraExpress FS DNA Library Prep Kit has been developed in response to user need for a faster, streamlined DNA prep workflow. This kit integrates enzymatic fragmentation and delivers high quality libraries from a variety of sample types. It features a single protocol for all DNA inputs, ranging from 10 – 200 ng of intact DNA. The workflow incorporates master mixed reagents, reduced incubation times, and fewer cleanup steps. Use of this kit also generates less plastic consumable waste because the entire library prep is conducted in a single tube.
- Go from intact sample to library in under 2 hours with FS (Fragmentation System) enzymatic fragmentation
- Single-protocol simplicity cuts down on reaction setup time, while streamlined workflows speed up library prep
- A single-tube workflow means less plastic and consumable waste
- Flexibility is enabled with simple guidelines for customized protocols, if desired
- Automation-friendly protocols for enhanced scalability
- 产品类别:
- Library Preparation for Illumina Products,
- DNA Library Prep for Illumina,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E3340S -20 NEBNext UltraExpress FS Enzyme Mix E3343AVIAL -20 1 x 0.024 ml Not Applicable NEBNext UltraExpress FS Reaction Buffer E3344AVIAL -20 1 x 0.096 ml Not Applicable NEBNext UltraExpress Ligation Master Mix E3345AVIAL -20 1 x 0.48 ml Not Applicable NEBNext MSTC High Yield Master Mix E3346AVIAL -20 1 x 1.08 ml Not Applicable TE Buffer (1X) E3347AVIAL -20 1 x 0.72 ml 1 X NEBNext Bead Reconstitution Buffer E3348AVIAL -20 1 x 1.92 ml Not Applicable
-
E3340L -20 NEBNext UltraExpress FS Enzyme Mix E3343AAVIAL -20 1 x 0.096 ml Not Applicable NEBNext UltraExpress FS Reaction Buffer E3344AAVIAL -20 1 x 0.384 ml Not Applicable NEBNext UltraExpress Ligation Master Mix E3345AAVIAL -20 2 x 0.96 ml Not Applicable NEBNext MSTC High Yield Master Mix E3346AAVIAL -20 1 x 4.32 ml Not Applicable TE Buffer (1X) E3347AAVIAL -20 1 x 2.88 ml 1 X NEBNext Bead Reconstitution Buffer E3348AAVIAL -20 1 x 7.68 ml 1 X
-
-
相关产品
相关产品
- T2030 Monarch RNA Cleanup Kit 10 ug
- NEBNext® 多样本接头引物试剂盒 5(96 种 Unique 双端 Index 引物)
- NEBNext® 文库定量试剂盒(Illumina®)
- NEBNext® 磁性分离架
操作说明、说明书 & 用法
-
操作说明
- Where can I find protocols for using the NEBNext UltraExpress™ FS DNA Library Prep Kit NEB #E3340)?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE3340
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- When preparing samples using the NEBNext UltraExpress™ FS DNA Library Prep Kit, can my input DNA be in EDTA-containing solutions?
- What if I see a precipitate in the NEBNext UltraExpress™ FS Reaction Buffer?
- Do I really need to vortex the NEBNext UltraExpress™ FS Enzyme Mix?
- The ligation reaction is very viscous. What will happen if it is not mixed properly?
- How much starting material must I use when preparing libraries with the NEBNext UltraExpress™ FS DNA Library Prep Kit?
- What sample types can I use with the NEBNext UltraExpress™ FS DNA Library Prep Kit?
- Can I use the NEBNext UltraExpress™ FS DNA Library Prep Kit for bisulfite conversion and EM-seq™ workflows?
- Is it possible to use sheared DNA as input material with the NEBNext UltraExpress™ FS DNA Library Prep Kit?
- What is the difference between the NEBNext UltraExpress™ FS DNA Library Prep Kit and the NEBNext® Ultra™ II FS DNA Library Prep Kit? How do I choose the right one?
- Do the NEBNext UltraExpress™ Library Prep kits come with beads?
- Which NEBNext® Multiplex Oligos (Adaptors & Primers) can be used with the NEBNext UltraExpress Library Prep Kits?
- What should I do if I see adaptor dimer in my NEBNext UltraExpress™ libraries?
- Can this kit be used with adaptors and primers from suppliers other than NEB?
- Can I use the NEBNext UltraExpress™ FS DNA Library Prep Kit to prepare libraries for sequencing on Ion Torrent instruments?
- Does the fragmentation reagent in NEBNext UltraExpress™ FS DNA kits contain NEBNext dsDNA Fragmentase®?
- Can the NEBNext UltraExpress™ FS DNA Library Prep Kit provide sufficient library yields for target enrichment?
