上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
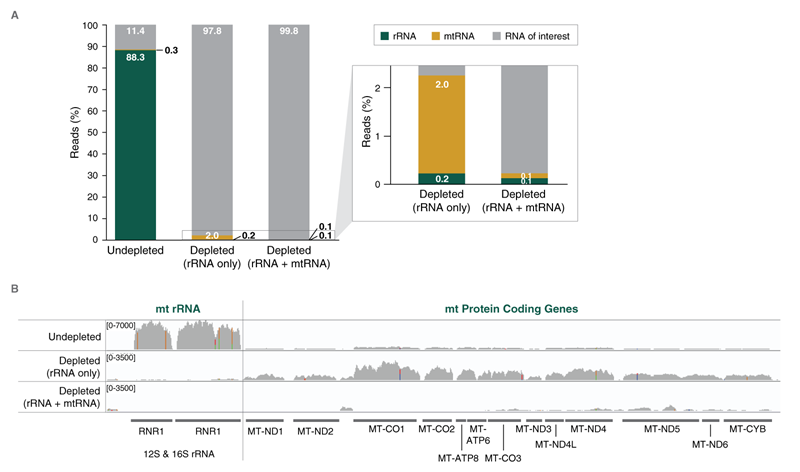
RNA-seq samples can include a large dynamic range of transcript expression, and unwanted, abundant RNAs must be removed in order to achieve efficient sequencing of the RNAs of interest. While pre-designed kits are available for some sample types (e.g. human, mouse, rat, some bacteria) and abundant RNAs (e.g. rRNA, globin mRNA), a customized approach is required for rRNA removal from other sample types as well as for removal of specific non-rRNA RNAs.
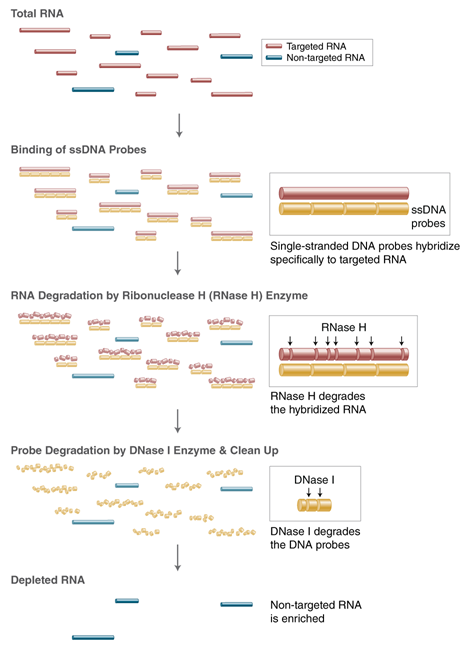
Customized depletion of unwanted RNAs is enabled by use of the NEBNext RNA Depletion Core Reagent together with custom probes, following this workflow:
STEP 1: Use the online NEBNext Custom RNA Depletion Design Tool to obtain custom probe sequences, by entering the sequence of your target RNA.
STEP 2: Order ssDNA probe oligonucleotides from your trusted oligo provider.
STEP 3: Use the probes with the NEBNext Custom RNA Depletion Core Reagent Set or in combination with other NEBNext RNA Depletion Kits.
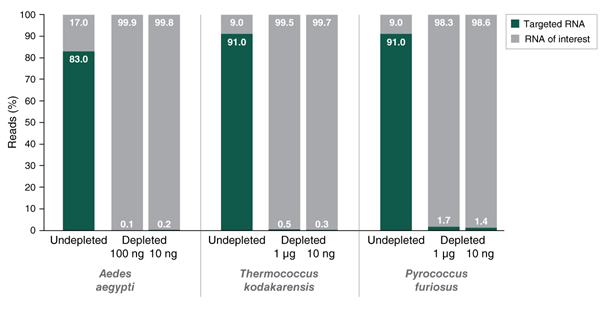
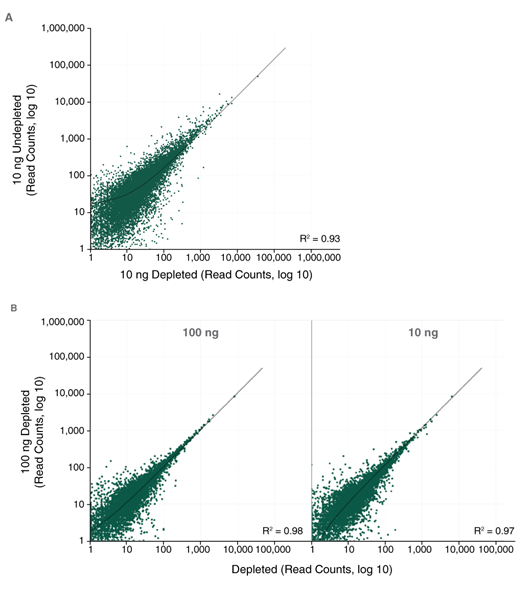
The NEBNext RNaseH-based RNA depletion workflow together with the closely-tiled custom probes designed using the online tool, allow effective depletion from both low- and high-quality RNA, with a broad range of input amounts.
The kit is also available with RNAClean® beads.
Features
- Customizable option to deplete unwanted RNA from any organism, using probe sequences designed with the NEBNext Custom RNA Depletion Design Tool.
- Compatible with a broad range of input amounts: 10 ng – 1 μg
- Suitable for low-quality or high-quality RNA
- Fast workflow: 2 hours, with less than 10 minutes hands-on time
- 产品类别:
- RNA Depletion & mRNA Enrichment Products,
- RNA Library Prep for Illumina,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E7865S -20 NEBNext® DNase I E7753-2VIAL -20 1 x 0.015 ml 20,000 units/ml NEBNext® Thermostable RNase H E7752-2VIAL -20 1 x 0.012 ml 5,000 units/ml RNase H Reaction Buffer E6312-2VIAL -20 1 x 0.012 ml Not Applicable DNase I Reaction Buffer E6315-2VIAL -20 1 x 0.03 ml Not Applicable Nuclease-free Water E6317-2VIAL -20 1 x 0.4 ml Not Applicable NEBNext Probe Hybridization Buffer E6314-2VIAL -20 1 x 0.012 ml Not Applicable
-
E7865L -20 NEBNext® DNase I E7753-3VIAL -20 1 x 0.06 ml 20,000 units/ml NEBNext® Thermostable RNase H E7752-3VIAL -20 1 x 0.048 ml 5,000 units/ml RNase H Reaction Buffer E6312-3VIAL -20 1 x 0.048 ml Not Applicable DNase I Reaction Buffer E6315-3VIAL -20 1 x 0.12 ml Not Applicable Nuclease-free Water E6317-3VIAL -20 1 x 1.5 ml Not Applicable NEBNext Probe Hybridization Buffer E6314-3VIAL -20 1 x 0.048 ml Not Applicable
-
E7865X -20 NEBNext® DNase I E7753-4VIAL -20 1 x 0.24 ml 20,000 units/ml NEBNext® Thermostable RNase H E7752-4VIAL -20 1 x 0.192 ml 5,000 units/ml RNase H Reaction Buffer E6312-4VIAL -20 1 x 0.192 ml Not Applicable DNase I Reaction Buffer E6315-4VIAL -20 1 x 0.48 ml Not Applicable Nuclease-free Water E6317-4VIAL -20 1 x 6 ml Not Applicable NEBNext Probe Hybridization Buffer E6314-4VIAL -20 1 x 0.192 ml Not Applicable
-
-
相关产品
相关产品
- e7760-nebnext-ultra-ii-directional-rna-library-prep-kit-for-illumina
- E7850 NEBNext rRNA Depletion Kit Bacteria
- E7400 NEBNext rRNA Depletion Kit v2 Human Mouse Rat
- E7405 NEBNext rRNA Depletion Kit v2 Human Mouse Rat with RNA Sample Purification Beads
- E7750 NEBNext Globin and rRNA Depletion Kit Human Mouse Rat
- E7755 NEBNext Globin and rRNA Depletion Kit Human Mouse Rat with RNA Sample Purification Beads
- NEBNext® Ultra™ II RNA 非链特异性文库制备试剂盒
操作说明、说明书 & 用法
-
操作说明
- Where can I find protocols for using NEBNext® RNA Library Prep reagents?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE7865_E7870
-
使用指南
- Guidelines for use of Custom RNA Depletion Probe Pools with the NEBNext RNA Depletion Core Reagent Set
工具 & 资源
-
Web 工具
- NEBNext Custom RNA Depletion Design Tool
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- How can I design DNA probes to use with the NEBNext RNA Depletion Core Reagent Set?
- Are the probes designed by the NEBNext Custom RNA Depletion Design Tool sense or antisense?
- Can the input sequence into the NEBNext Custom RNA Depletion Design Tool contain IUPAC Ambiguity Code?
- Does the NEBNext Custom RNA Depletion Design Tool check for probes targeting transcripts other than the ones provided?
- I have used NEBNext Custom RNA Depletion Design Tool and obtained probe sequences, what’s the next step?
- How do I make a working probe pool to be used with the NEBNext RNA Depletion Core Reagent Set?
- What’s the maximum number of probes that can be combined in a pool?
- Can I use this product with degraded RNA or fragmented RNA?
- What total RNA input should I use with the NEBNext RNA Depletion Core Reagent Set?
- Why must the RNA be free of DNA?
- How can I determine if the RNA depletion was efficient?
- Can I use Agencourt®AMPure XP Magnetic Beads instead of RNAClean® XP Magnetic Beads/NEBNext® RNA Sample Purification Beads?
- Can I use purification methods other than NEBNext® RNA Sample Purification Beads/Agencourt® RNAClean® XP Magnetic Beads?
- Does the NEBNext Custom RNA Depletion Tool check for probe redundancy or overlap between target RNA sequences entered?
- Can the enriched RNA resulting from the NEBNext depletion kit be used in long read sequencing applications?
-
问题解决指南
- Troubleshooting Guide for use with the NEBNext® RNA Depletion Core Reagent Set and NEBNext Custom RNA Depletion Tool
